Abstract
In humans and many other animals, memory consolidation occurs through multiple temporal phases and usually involves more than one neuroanatomical brain system. Genetic dissection of Pavlovian olfactory learning in Drosophila melanogaster has revealed multiple memory phases, but the predominant view holds that all memory phases occur in mushroom body neurons. Here, we demonstrate an acute requirement for NMDA receptors (NMDARs) outside of the mushroom body during long-term memory (LTM) consolidation. Targeted dsRNA-mediated silencing of Nmdar1 and Nmdar2 (also known as dNR1 or dNR2, respectively) in cholinergic R4m-subtype large-field neurons of the ellipsoid body specifically disrupted LTM consolidation, but not retrieval. Similar silencing of functional NMDARs in the mushroom body disrupted an earlier memory phase, leaving LTM intact. Our results clearly establish an anatomical site outside of the mushroom body involved with LTM consolidation, thus revealing both a distributed brain system subserving olfactory memory formation and the existence of a system-level memory consolidation in Drosophila.
Pavlovian olfactory learning in Drosophila creates an elemental associative memory in a simple, accessible insect brain. This form of behavioral plasticity requires the normal function of NMDARs1, as is the case in other invertebrate and vertebrate species2,3. Memory formation in flies thereafter proceeds through several temporal phases and involves multiple biochemical cascades, which is again similar to findings from other animal models4–6. In particular, spaced training produces stronger, longer-lasting memory than massed training, which is a common property of memory formation7,8. Contrary to the fact that memory storage involves multiple anatomical regions in other animal models9, olfactory memory has been proposed to be stored predominantly in the mushroom body neurons5,10.
The mushroom body is a prominent neuropillar structure in the insect central brain. Intrinsic mushroom body cells send neurites ventrally into the calyx, a region of dendritic arborization that is innervated by efferents from several different regions, including projection neurons from the antennal lobes11. Mushroom body axons project rostrally as a densely packed and stalk-like structure called the pedunculus to the anterior face of the brain, where they split and give rise to the dorsally projecting α and α′ lobes and the medially projecting β, β′ and γ lobes12. Output neurons from the mushroom body project to many parts of the central brain. Another prominent neuropil is the central complex, which consists of four substructures, the ellipsoid body, the fan-shaped body, the nodulii and the protocerebral bridge. The central complex lies in the central brain between the pedunculi of the mushroom body and is bounded laterally by the two antennoglomerular tracts, dorsally by the pars intercerebralis, ventrally by the esophagus and the great commissure and frontally by the median bundle and the β-lobes of the mushroom bodies frontally13. The central complex forms intricate connections to a variety of brain centers, may mediate communication between the two hemispheres and is believed to be a control center for many different behavioral outputs13,14.
The mushroom body does indeed have a central role in Pavlovian olfactory learning5,6,10. Nevertheless, emerging evidence has hinted at the involvement of other extrinsic neurons and anatomical sites during olfactory memory consolidation. The cer gene, encoding a cathepsin inhibitor, most likely regulates LTM through its expression in glial cells surrounding the mushroom body15. In addition, several enhancer-trap transposon insertions with expression patterns exclusively outside of the mushroom body have been identified in a behavioral screen for olfactory memory mutants, although the functional relevance of these enhancer traps remains to be verified16,17. Finally, an ‘asymmetrical structure’ in the central complex seems to correlate with the presence of LTM, but the functional relevance of this observation is also unknown18.
We recently have shown that NMDARs function in Drosophila during olfactory learning and thereafter during LTM consolidation1. Here, using newly-constructed, dsRNA-mediated, UAS-driven transgenes to silence dNR1 or dNR2, we have disrupted the normal function of NMDARs in the ellipsoid body or in the mushroom body, and have silenced synaptic transmission from both structures with UAS-driven dominant-negative UAS-shits1 (ref. 19). We demonstrate dual dissociable roles for NMDARs in the ellipsoid body during LTM consolidation and in the mushroom body during the middle-term memory (MTM) phase of early memory processing. Our results suggest that LTM is stored in the ellipsoid body, outside of the mushroom body.
RESULTS
LTM is abolished by disrupting NMDARs in the ellipsoid body
Functional NMDARs in Drosophila consist of two subunits, dNR1 and dNR2, both of which are expressed widely in the adult brain, including in the mushroom body1 and central complex (Supplementary Figs. 1 and 2 online). To address where functional NMDARs are required during olfactory memory formation, we used the binary GAL4/UAS gene-expression system20 to target dsRNA-mediated knockdown of either the dNR1 or dNR2 subunits to various brain regions. For this purpose, we generated dsRNA-based transgenes (UAS-dsNR1 and UAS-dsNR2) to silence gene expression in a sequence-specific manner21. dsRNA triggers RNAi interference, an evolutionarily conserved process of sequence-specific post-transcriptional gene silencing22,23. RNAi, however, can lead to the degradation of nontargeted mRNAs, commonly referred to as off-target effects. Using a recently established algorithm24 and homology blast, we identified zero legitimate off-target sequences for UAS-dsNR1 and only one (CG17124) for UAS-dsNR2 (Supplementary Table 1 online). Therefore, dsRNA-mediated effects from both transgenes appeared to be highly specific for dNR1 and dNR2, and thus to functional NMDARs. Consistent with this expectation, the UAS-dsNR1 transgene disrupted LTM in a quantitatively and qualitatively similar manner (Supplementary Fig. 3 online), as we previously have shown with a specific dNR1 antisense message1. We have not yet constructed a second dsRNA transgene for dNR2. dNR1 and dNR2 function together in Drosophila1, however, so the silencing of dNR2 constitutes a third disruption of NMDAR function. The fact that the LTM defect from our UAS-dsNR2 transgene is similar to those from genetic disruptions of dNR1 (see below) provides strong in vivo evidence against any different off-target effects from these transgenes. Taken together, the combined use of UAS-dsNR1 and UAS-dsNR2 provided us with a specific dsRNA-mediated silencing of functional NMDARs.
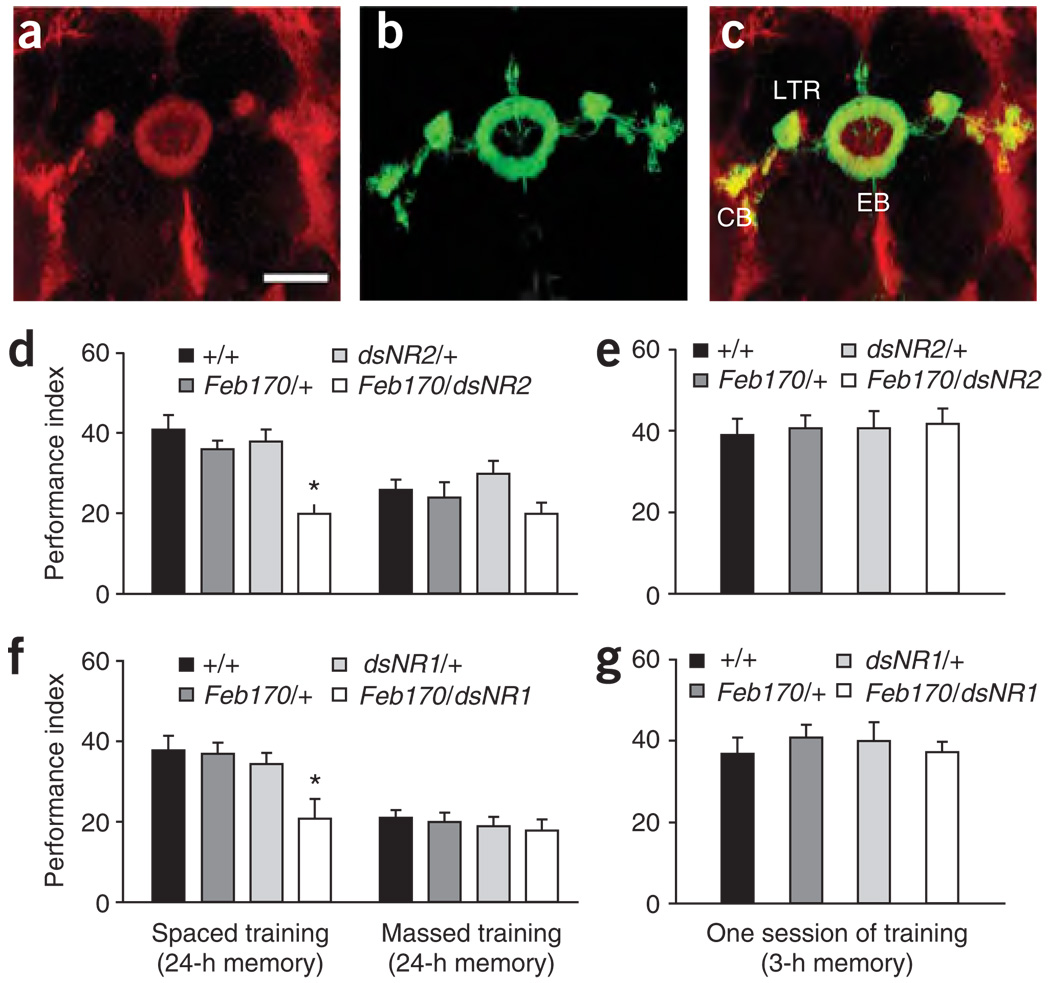
Western analysis revealed that the dNR2 protein was barely detectable in C155/+; UAS-dsNR2/+ flies (elav/dsNR2), whereas the heterozygous C155 (elav/+) and UAS-dsNR2 (dsNR2/+) control flies produced a clear, single band that corresponded to the predicted sizes of the dNR2-2 and dNR2-3 proteins (Fig. 1a and Supplementary Fig. 2). Silencing of dNR2 with UAS-dsNR2 was further confirmed in situ with immunohistochemistry. Including our previously published antibody to dNR2 (monoclonal α-5B10), we have developed four antibodies to dNR2 that could be used for immunohistochemistry (see Supplementary Figs. 1 and 2 for details). All four antibodies labeled the ellipsoid body, suggesting that dNR2 was expressed preferentially in the ellipsoid body. Notably, two of these antibodies (α-820-2 and α-820-1) strongly labeled the ellipsoid body, suggesting an elevated expression of dNR2 in the ellipsoid body (Fig. 1b and Supplementary Fig. 2). This elevated expression was specifically diminished in Feb170/+; UAS-dsNR2/+ flies (Fig. 1c,d; see below for characterization of Feb170), whereas the expression of dNR2 in the protocerebral bridge (Fig. 1c,d) or in the optical lobes (Fig. 1e,f) was not affected. This reduction in dNR2 did not affect the gross morphology of the ellipsoid body, however, suggesting that a knockdown of dNR2 to this degree is not necessary for normal development of the ellipsoid body (Fig. 1g,h).
Figure 1.
dsRNA-mediated knockdown of dNR2 in adult brain. (a) A schematic of the UAS-dsNR2 transgenic construct is shown (left panel; see Methods for details). Western blot analysis revealed an appreciable knockdown of the dNR2 protein in adult heads of C155/+; UAS-dsNR2/+ flies (elav/dsNR2) compared with C155 (elav/+) and UAS-dsNR2 (dsNR2/+) control genotypes (right panel). (b,c) Immunostaining for dNR2 was reduced in R4m neurons of the ellipsoid body in Feb170/+; UAS-dsNR2/+ flies (Feb170; dsNR2, c) compared with Feb170/+ control flies (Feb170, b). (d) Immunostaining of dNR2 was quantified in R4m neurons of the ellipsoid body and in the nearby protocerebral bridge for Feb170/+ (black bars) and Feb170/+; UAS-dsNR2/+ (white bars) flies. (e,f) Immunostaining of dNR2 was similar in optical lobes of Feb170/+ (Feb170; e) and Feb170/+; UAS-dsNR2/+ (Feb170; dsNR2; f) flies. (g,h) In spite of the knockdown of dNR2 in Feb170/+; UAS-dsNR2/+ flies (see above), the gross morphology of the ellipsoid body was normal in Feb170/+; UAS-dsNR2, UAS-mCD8::GFP/+ flies (Feb170; GFP, dsNR2, g), as compared to Feb170/+; UAS-mCD8::GFP/+ flies (Feb170; GFP, h). Scale bar, 50 µm.
Western analysis also indicated that the dNR1 protein was reduced greatly in UAS-dsNR1/+; hs-GAL4/+ flies (P26/dsNR1) after heat shock, as compared with the heterozygous hs-GAL4 (P26/+) or UAS-dsNR1 (dsNR1/+) control flies (Supplementary Fig. 3). This confirmed our previous report that dNR1 could be acutely disrupted with heat shock using a heat shock–inducible GAL4 driver (P26)1. Behavioral analyses suggested that LTM was abolished, but anesthesia-resistant memory (ARM) was normal in UAS-dsNR1/+; hs-GAL4/+ flies after heat shock–induced knockdown of dNR1 (Supplementary Fig. 3), confirming our previous report using a different genetic method to knockdown dNR1.
The strong expression of dNR2 in ellipsoid body interested us, as another closely-related substructure of the central complex, the fan-shaped body, appears to subserve a short-term memory trace for visual learning25 and may be involved with LTM formation after courtship conditioning26. We identified several GAL4 drivers with preferential expression in these dNR2-positive ellipsoid body neurons (Fig. 2 and Supplementary Figs. 4 and 5 online). Three GAL4 drivers, Feb170 (ref. 27), Ruslan (ref. 16) and C42 (ref. 28), were chosen for further analyses, as their expression patterns overlapped only in these NR2-positive neurons of the ellipsoid body (Supplementary Fig. 5). Notably, all threeGAL4 drivers showed no expression in the mushroom body (Supplementary Fig. 5). The ellipsoid body contains ten types of small-field neurons that connect it to other central complex substructures (for example, the fan-shaped body)13. It also contains at least six types of large-field (R) neurons that arborize as a ring, mainly in the anterior part13,28. Genetic mosaic flip-out analyses and immunolabeling indicated that the dNR2-positive neurons are the large-field, R4m subtype (H.-H. Lin, and A.-S.C., unpublished data). These dNR2-positive R4m neurons are likely cholinergic, as indicated by Cha-GAL80 inhibition of their GAL4 expression (Supplementary Fig. 5).
Figure 2.
Functional NMDARs in the ellipsoid body are required for LTM. (a–c) Preferential expression of dNR2 and Feb170 GAL4 in R4m neurons of the ellipsoid body. Confocal imaging of dNR2 immunostaining (with α-820-2) in whole-mount adult brain revealed preferential expression (red) in R4m neurons of the ellipsoid body (posterior view, a). The Feb170 GAL4-driven expression of a UAS-mCD8::GFP reporter (green) in the ellipsoid body is shown (b). Merged images indicated overlap (yellow) of dNR2 staining (red) and GFP reporter (green) in R4m neurons of the ellipsoid body (EB, c). CB, cell body; LTR, lateral triangle. Scale bar, 50 µm. (d) dsRNA-mediated knockdown of dNR2 via Feb170 GAL4-driven expression of a UAS-dsNR2 transgene specifically abolished LTM. We observed that 1-d memory after spaced training was significantly disrupted (* P < 0.05), whereas 1-d memory after massed training was normal in Feb170/+; UAS-dsNR2/+ (Feb170/dsNR2) flies. Heterozygous Feb170 (Feb170/+) and UAS-dsNR2 (dsNR2/+) control flies produced similar scores to wild-type flies (+/+) after either spaced or massed training (n = 8 performance indexes per group). (e) dsRNA-mediated knockdown of dNR2 via Feb170 GAL4-driven expression of the same UAS-dsNR2 transgene had no effect on 3-h memory after one session of training in Feb170/+; UAS-dsNR2/+ (Feb170/dsNR2) flies, as compared to control flies (+/+, Feb170/+ or dsNR2/+) (n = 8 performance indexes per group). (f) dsRNA-mediated knockdown of dNR1 via Feb170 GAL4-driven expression of a UAS-dsNR1 transgene specifically abolished LTM, similar to knockdown of dNR2 (n = 8 performance indexes per group). (g) dsRNA-mediated knockdown of dNR1 via Feb170 GAL4-driven expression of a UAS-dsNR1 transgene had no effect on 3-h memory after one session of training, similar again to knockdown of dNR2 (n = 8 performance indexes per group).
Because we focused on genetic manipulations of dNR1 in our previous study1, we first disrupted the functional NMDARs in the ellipsoid body by knockdown of dNR2, and then confirmed our results with similar genetic manipulations of dNR1 (which also is expressed in the ellipsoid body; ref. 1 and Supplementary Fig. 2). Knockdown of dNR2 using Feb170-GAL4 to drive UAS-dsNR2 specifically reduced the dNR2 protein expression in the ellipsoid body (Fig. 1b–f), without affecting the gross morphology of the ellipsoid body (Fig. 1g,h). The same knockdown also produced a 50% reduction in 1-d memory after spaced training (Fig. 2d). Similarly, disruption of NMDARs by Feb170-driven transgenic expression of UAS-dsNR1 specifically disrupted 1-d memory after spaced training (Fig. 2f). We further confirmed this disruptive effect on 1-d memory after spaced training with Ruslan and C42 to drive UAS-dsNR2 in the ellipsoid body (Supplementary Fig. 4). In contrast, the same genetic manipulations had no effects on 1-d memory after massed training (right panels, Fig. 2d,f), Feb170-driven expression of UAS-dsNR2 (Fig. 2e) or UAS-dsNR1 (Fig. 2g) had no effect on 3-h memory after one session of training and Feb170-driven expression of UAS-dsNR2 (Supplementary Fig. 6 online) or UAS-dsNR1 (Supplementary Fig. 6) had no effect on memory immediately after one session of training. Together with other behavior-genetic dissections dividing olfactory memory formation into four functionally distinct memory phases—short-term memory (STM), MTM, ARM and LTM8,17,29—these observations suggest that genetic disruption of NMDAR function in the ellipsoid body specifically abolishes LTM, without affecting other earlier memory phases.
Protein synthesis–dependent LTM is specifically abolished
To rule out the possibility that a developmental defect underlay this LTM defect, we used the tubulin (tub)-GAL80ts transgene with the binary GAL4/UAS system30. Groups of flies, raised at 18 °C, were kept for 3 d at either 18 °C or 29 °C before spaced training. We determined that incubation for 3 d at 29 °C was sufficient for optimal disinhibition of a Feb170-driven UAS-mCD8::GFP reporter by tub-GAL80ts (Supplementary Fig. 7 online). We observed that 1-d memory after spaced training was reduced significantly in transgenic Feb170/+; UAS-dsNR2/tub-GAL80ts flies when kept at 29 °C for 3 d, whereas such memory was normal in the same flies when kept at 18 °C (P o 0.05; Fig. 3a), suggesting an acute physiological role for functional NMDARs in the ellipsoid body during LTM processing.
Figure 3.
Inducible knockdown of dNR2 specifically blocks consolidation and storage, but not retrieval, of protein synthesis–dependent LTM. (a) Adult-specific knockdown of dNR2 using the GAL80ts repressor of GAL4-mediated UAS-dsNR2 expression specifically abolished LTM at the restrictive temperature. After spaced training, 1-d memory was significantly disrupted in Feb170/+; UAS-dsNR2/+; tub-GAL80ts/+ (Feb170/dsNR2; GAL80ts) flies when they were shifted from 18 °C to 29 °C for 3 d (right panel; n = 16 performance indexes per group) before training (SP, red arrow) and tested (black arrow) at 29 °C, but was normal when they were kept at 18 °C throughout the experiment (left panel; n = 8 performance indexes per group). (b) Adult-specific knockdown of dNR2 abolished protein synthesis–dependent LTM. After spaced training, 1-d memory was reduced by 50% in CXM-fed wild-type flies (+/+ +CXM; left panel), but was not disrupted further in CXM-fed Feb170/+; UAS-dsNR2/+; tub-GAL80ts/+ (Feb170/dsNR2; GAL80ts +CXM) flies, kept for 3 d at 29 °C before training and testing (n = 8 performance indexes per group). (c) After spaced training, 1-d memory was normal in Feb170/+; UAS-dsNR2/+; tub-GAL80ts/+ (Feb170/dsNR2; GAL80ts) flies after 4 d at 29 °C, shifted to 18 °C for 3 d and followed by training and testing at 18 °C, indicating a recovery of NMDAR function at the permissive temperature. (d) Four-day memory after spaced training was significantly impaired in the same flies (Feb170/dsNR2; GAL80ts) when incubated at 29 °C for 3 d, subjected to spaced training, maintained at 29 °C for 1 d and then shifted back to 18 °C for 3 d before testing, indicating that knockdown of NMDARs for 1 additional day after training is sufficient to block LTM consolidation and storage. (e) After reversal spaced training, 1-d memory (RS, blue arrow) was higher in the same flies (Feb170/dsNR2; GAL80ts) than in controls when incubated at 29 °C for 3 d, subjected to spaced training, maintained at 29 °C for 1 d, shifted to 18 °C for 3 d, subjected to reversal training and tested at 18 °C 1 d later, indicating normal retrieval for the former (n = 8 performance indexes per group). * P < 0.05.
LTM uniquely depends on protein synthesis. Similar to previous studies8,29, 1-d memory was reduced by 50% in normal flies kept at 29 °C for 3 d and fed cycloheximide (CXM) overnight before spaced training (left, Fig. 3b). However, in transgenic Feb170/+; UAS-dsNR2/tub-GAL80ts flies kept at 29 °C for 3 d, CXM feeding did not further disrupt 1-d memory after spaced training (right, Fig. 3b). This result confirms that disruption of NMDARs specifically impairs protein synthesis–dependent LTM in the ellipsoid body.
Consolidation of LTM requires functional NMDARs
When kept at 29 °C (restrictive) for 4 d, shifted to 18 °C (permissive for inhibition of GAL4 by GAL80) for 3 d, subjected to spaced training and tested for 1-d memory at 18 °C, Feb170/+; UAS-dsNR2/tub-GAL80ts flies showed normal 1-d memory (Fig. 3c). This result suggests that adult-specific disruption of dNR2 during the 4-d incubation at 29 °C was restored to a normal functional level when the flies were shifted to 18 °C for 3 d. If flies were subjected to spaced training on day 3 of a 4-d incubation at 29 °C, however, 4-d memory (again at 18 °C for 3 d) was severely disrupted in Feb170/+; UAS-dsNR2/tub-GAL80ts flies (Fig. 3d). The dNR2 expression was restored to a normal functional level at the time of testing (Fig. 3c), thereby revealing a disruption of consolidation and storage, rather than of retrieval, of LTM by knockdown of dNR2 in Feb170/+; UAS-dsNR2/tub-GAL80ts flies.
A ‘reversal’ training protocol was designed to further distinguish a consolidation and storage defect from a retrieval failure. We subjected flies to a second reversal spaced training session 4 d after the first spaced training, where the original CS− became the CS+ (nonshocked odor and shocked odor, respectively), and vice versa. Thereafter, 1-d memory was quantified (performance indexes were calculated relative to the second CS+). In the absence of memory formation after the first spaced training session, reversal memory would be expected to be near a performance index of 35. If, on the other hand, memory formation after the first spaced training was normal, then it would counteract memory formation after reversal training, thereby producing a lower performance index that would be near zero. When UAS-dsNR2/tub-GAL80ts and Feb170/+; UAS-dsNR2/tub-GAL80ts flies were subjected first to spaced training on day 3 of a 4-d incubation at 29 °C and then to reversal spaced training after 3 d at 18 °C, 1-d reversal memory was significantly higher in Feb170/+; UAS-dsNR2/tub-GAL80ts flies than in UAS-dsNR2/tub-GAL80ts controls (Fig. 3e). This result demonstrates normal memory retrieval and implies weaker-than-normal memory storage from the first spaced training in Feb170/+; UAS-dsNR2/tub-GAL80ts flies. Therefore, functional NMDARs contribute specifically to the consolidation and storage, but not to the retrieval of a protein synthesis–dependent LTM in the ellipsoid body.
Overexpression of dNR2 in the ellipsoid body enhances LTM
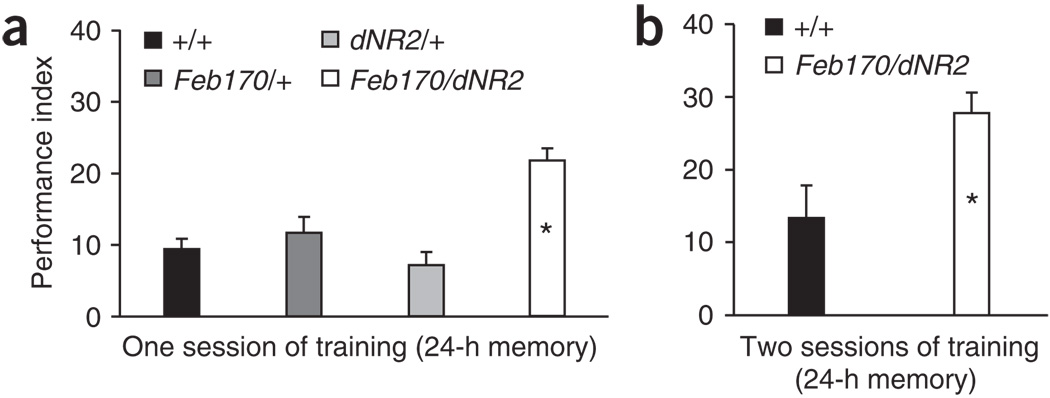
We observed that 1-d memory after one or two sessions of training was significantly enhanced when dNR2 was overexpressed in Feb170-targeted brain regions (Fig. 4a,b). This outcome is fully complementary to our dsRNA-based experiments: gene disruptions produce loss-of-function effects, whereas overexpression of the normal gene produces gain-of-function effects. Thus, NMDA receptors in Drosophila function in a manner that is quite analogous to a previous study in mice31.
Figure 4.
Overexpression of dNR2 in the ellipsoid body enhances 1-d memory. Males homozygous for UAS-dNR2-2 (a transgene expressing one of three dNR2 isoforms) were crossed with Feb170 or wild-type (+/+) virgins, and wild-type males were also crossed with Feb170 females. (a) We observed significantly higher 1-d memory after one session of training in Feb170/+; UAS-dNR2-2/+ flies (Feb170/dNR2) than in wild-type (+/+) flies or in Feb170/+ (Feb170/+) or UAS-dNR2-2/+ (dNR2/+) control flies (n = 18 performance indexes per group). (b) Similarly, 1-d memory after two sessions of training was also higher in Feb170/+; UAS-dNR2-2/+ flies (Feb170/dNR2) than in wild-type (+/+) controls (n = 12 performance indexes per group).
LTM retrieval requires neural activity from the ellipsoid body
A physiological requirement for NMDARs in the ellipsoid body during LTM consolidation argues that a newly acquired olfactory experience is initially processed in or upstream of the mushroom body5,6,10, but is then transferred from the mushroom body to the ellipsoid body for LTM storage. Such a hypothesis predicts that blocking synaptic output from the mushroom body, but not from the ellipsoid body, during memory consolidation (that is, during training and in a time window after training) would abolish LTM, and that blocking synaptic output from the ellipsoid body after memory consolidation is complete would disrupt LTM retrieval. To test these two predictions, we combined the UAS-shits1 transgene with Feb170 or OK107 GAL4 drivers. OK107 is a GAL4 driver targeting transgenic expression in all lobes of the mushroom body (Supplementary Fig. 5). The UAS-shits1 transgene has been shown to block neuronal transmission in a temperature-dependent, dominant-negative fashion19.
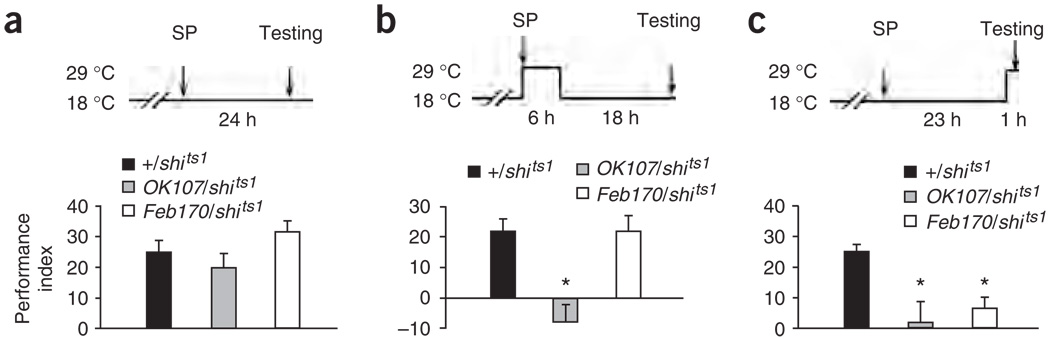
To test the first prediction, we subjected OK107/+; UAS-shits1/+ flies, Feb170/+; UAS-shits1/+ flies or UAS-shits1/+ controls to spaced training and then maintained them for the first 6 h thereafter at a restrictive temperature (29 °C). Compared with the results from control experiments carried out at 18 °C (Fig. 5a) or with control flies, 1-d memory after spaced training was abolished in OK107/+; UAS-shits1/+ flies, but was normal in Feb170/+; UAS-shits1/+ flies (Fig. 5b). To rule out the possibility that the 9-h blocking may cause irreversible disruption of synaptic transmission from the mushroom body, OK107/+; UAS-shits1/+ and UAS-shits1/+ flies were kept at 29 °C for 9 h and then shifted back to 18 °C for 18 h before being tested for initial learning after one session of training. Compared to UAS-shits1/+ controls, learning was normal in OK107/+; UAS-shits1/+ flies (68 ± 3.3 versus 61 ± 2.3; P = 0.1). This observation suggests that the abolition of 1-d memory in OK107/+; UAS-shits1/+ flies is specific to memory consolidation. These results demonstrate that neural activity from the mushroom body, but not from the ellipsoid body, is required during memory consolidation.
Figure 5.
Transference of memory from the mushroom body to the ellipsoid body during LTM consolidation. Wild-type (+/+), Feb170 or OK107 males were crossed to UAS-shits1 females, and all of the progeny were raised at 18 °C to minimize any potential leaky effect of UAS-shits1 on development. (a) Flies were kept at a permissive temperature (18 °C) throughout the entire experiment. After spaced training (SP), 1-d memory did not differ among UAS-shits1/+ (+/shits1), UAS-shits1/+; OK107/+ (OK107/shits1) or Feb170/+; UAS-shits1/+ (Feb170/shits1) flies. (b) Flies were shifted to a restrictive temperature (29 °C), subjected to spaced training, maintained at 29 °C for another 6 h, shifted back to 18 °C for 18 h and tested at 18 °C. We observed that 1-d memory after spaced training was abolished in UAS-shits1/+; OK107/+ flies, but not in Feb170/+; UAS-shits1/+ flies, compared with UAS-shits1/+ controls. (c) Flies were subjected to spaced training at 18 °C, kept at 18 °C for 23 more h, shifted to 29 °C and then tested 1 h later. UAS-shits1/+; OK107/+ andFeb170/+; UAS-shits1/+ flies had impaired 1-d memory after spaced training, compared with UAS-shits1/+ controls, indicating that the output of neural activity from both the mushroom body and ellipsoid body is required during retrieval of LTM. In each of the above experiments, wild-type flies were also tested under the same conditions and showed consistent, higher scores (performance index = 48 ± 3, 40 ± 2, and 48 ± 5 for experiments a–c) than UAS-shits1/+ flies, as reported by others39. n = 8 performance indexes per group.
To test the second prediction, we subjected the same three genotypes to spaced training at 18 °C, maintained them at 18 °C for 23 h after training and then shifted them to 29 °C for 1 h before testing them at 29 °C. Under these conditions, 1-d memory after spaced training was blocked in both OK107/+; UAS-shits1/+ and Feb170/+; UAS-shits1/+ flies (Fig. 5c). Under the same conditions, however, sensorimotor responses to the odors were normal in both OK107/+; UAS-shits1/+ and Feb170/+; UAS-shits1/+ flies, as compared to UAS-shits1/+ controls (48 ± 4.1 or 40 ± 5.3 versus 49 ± 5.1, P = 0.34 for 3-octanol; 48 ± 3.3 or 39 ± 4.9 versus 39 ± 5.1, P = 0.28 for 4-methyl-cyclohexanol). This observation suggests that memory retrieval is specifically disrupted in both OK107/+; UAS-shits1/+ and Feb170/+; UAS-shits1/+ flies. Therefore, after LTM is consolidated, neural activity from either the mushroom body or ellipsoid body is required for retrieval. Together, these observations argue that consolidation and storage of olfactory LTM occurs upstream of synaptic transmission from the R4m neurons of the ellipsoid body. Considering that memory may be transferred outside of the mushroom body (during and within the first 6 h after training) for LTM consolidation (Fig. 5b) and that NMDARs are specifically required in the ellipsoid body for such a process (Figs. 2–4), our results suggest that early memory resides in (or upstream of) the mushroom body and then is transferred to the ellipsoid body during the consolidation of LTM.
NMDARs in the mushroom body are not required for LTM
The specific requirement of functional NMDARs in the ellipsoid body for LTM consolidation and storage prompted us to explore the role of NMDARs in the mushroom body (Fig. 6), the neuroanatomical structure known to mediate olfactory associations, STM and probably LTM5,10. dNR2 or dNR1 in the mushroom body were specifically diminished when UAS-dsNR2 (Fig. 6a–d) or UAS-dsNR1 (Fig. 6g–j) were targeted to the mushroom body with OK107, whereas their expression in the optical lobes were not affected. We observed that 1-d memory after spaced or massed training was normal when dNR2 (Fig. 6e) or dNR1 (Fig. 6k) was knocked down, suggesting that functional NMDARs in the mushroom body are not required for the formation of LTM or ARM.
Figure 6.
NMDAR function in the mushroom body is required for MTM but not for LTM. (a,b) Immunostaining of dNR2 was specifically reduced in the mushroom body calyx (dendrites) in OK107/+; UAS-dsNR2/+ flies (dsNR2; OK107, b), as compared with OK107/+ controls (OK107, a). (c,d) dNR2 expression in optical lobes was similar between these flies. Scale bar, 50 µm. (e) dsRNA-mediated knockdown of dNR2 in the mushroom body did not affect 1-d memory after spaced or massed training in OK107/dsNR2 flies, compared to controls (+/+, OK107/+ or dsNR2/+), indicating that NMDAR function in the mushroom body is not required for LTM. (f) dsRNA-mediated knockdown of dNR2 in the mushroom body disrupted 3-h memory after one session of training (without cold shock) in OK107/dsNR2 flies, compared with control (+/+, OK107/+ or dsNR2/+) flies. In contrast, 3-h memory after one session of training did not differ among these genotypes if the flies were subjected to a 2-min cold shock 2 h after training. Together, these data indicate that knockdown of dNR2 in the mushroom body impairs MTM, but not ARM8 or LTM. (g–j) Similarly, immunostaining of dNR1 was specifically reduced in the mushroom body calyx in UAS-dsNR1/+; OK107/+ flies (dsNR1; OK107, h), as compared with OK107/+ controls (g), but was not affected in optical lobes in the same flies (i,j). (k) dsRNA-mediated knockdown of dNR1 via OK107 GAL4–driven expression of the same UAS-dsNR1 transgene did not affect 1-d memory after spaced or massed training, similar to knockdown of dNR2. (l) dsRNA-mediated knockdown of dNR1 via OK107 GAL4–driven expression of the UAS-dsNR1 transgene impaired MTM, but not ARM, again as with knockdown of dNR2. n = 8 performance indexes per group for 1-day memory and n = 6 performance indexes per group for 3-h memory.
The latter conclusion was further confirmed by assessing 3-h memory after one session of training, which can be broken down into an anesthesia-sensitive memory (ASM) and ARM8. Knockdown of dNR2 (Fig. 6f) or dNR1 (Fig. 6l) via the OK107 mushroom body GAL4 driver similarly disrupted 3-h memory after one session of training. In contrast, 3-h memory was normal in these same transgenic flies when subjected to a 2-min cold shock 2 h after training, which provides a direct measure of ARM8. Similar effects were also observed when UAS-dsNR1 was targeted to the mushroom body with two additional mushroom body GAL4 drivers, C739 and 201Y (Supplementary Figs. 5 and 8 online). Together, these data indicate that functional NMDARs in the mushroom body contribute to ASM. Memory retention immediately after one session of training was normal in transgenic flies expressing UAS-dsNR2 or UAS-dsNR1 via the OK107 mushroom body GAL4 driver (Supplementary Fig. 6), suggesting that STM is also not affected by knockdown of NMDARs in the mushroom body. Therefore, MTM, but not STM, is the component memory phase of ASM that was specifically disrupted.
OK107 is a GAL4 driver labeling almost all of the mushroom body neurons32. Some of these neurons were probably cholinergic, including the γ lobe neurons that are primarily targeted by 201Y and a subset of the α/β lobe neurons that are primarily labeled by C739 (Supplementary Fig. 5). The rest, a subset of the α/β lobe neurons, were likely noncholinergic (Supplementary Fig. 5). Therefore, the cholinergic γ lobe neurons are targeted by both OK107 and 201Y, whereas the cholinergic α/β lobe neurons are targeted by bothOK107 and C739. We observed that 3-h memory without cold shock in OK107/dsNR2 flies did not differ from that with cold shock in control flies (Fig. 6f), suggesting that MTM was completely abolished. In contrast, MTM was only partially disrupted in 201Y/dsNR1 or in C739/dsNR1 flies (Supplementary Fig. 8). These results argue that MTM depends on cholinergic transmission and is completely abolished when NMDARs are silenced in all of these cholinergic α/β and γ cells.
DISCUSSION
The predominant view of olfactory memory is that it is processed in the mushroom body5,10. Our data demonstrate a role for R4m neurons in the ellipsoid body during LTM consolidation, which now suggests a much broader and more complex neuronal circuitry sub-serving olfactory memory consolidation in Drosophila. Because genetic modulations of NMDARs in the ellipsoid body produce effects that are specific to LTM, components of this complex neural circuitry appear to subserve at least one specific temporal phase (LTM) of memory processing. Consistent and complementary to this notion, consolidation of LTM remains normal when NMDAR function is disrupted in the mushroom body (Table 1). Thus, the transference of memory from one anatomical location to another as consolidation progresses to LTM appears to occur in Drosophila, as in various other species6,9,33.
Table 1.
Effects of specific disruption of functional NMDARs in ellipsoid body or mushroom body on four different memory phases after olfactory learning
| Disruption of NMDARs in ellipsoid body (with Feb170 as the GAL4 driver) |
Disruption of NMDARs in mushroom body (with OK107 as the GAL4 driver) |
|||
|---|---|---|---|---|
| UAS-dsNR2 | UAS-dsNR1 | UAS-dsNR2 | UAS-dsNR1 | |
| LTM | ++ | ++ | − | − |
| MTM | − | − | ++ | + |
| ARM | − | − | − | − |
| STM | − | − | − | − |
++ indicates complete abolition; +, severe disruption; −, no effect at all.
NMDARs in the ellipsoid body and LTM consolidation
Specific disruption of functional NMDARs by dsRNA-mediated knockdown of either dNR2 or dNR1 in the R4m neurons of the ellipsoid body specifically abolishes protein synthesis–dependent LTM (Figs. 2 and 3 and Table 1), suggesting that the ellipsoid body is important during memory consolidation. NMDAR function is physiological rather than developmental, as induction of dsRNA transgenes in adults was sufficient to abolish LTM (Fig. 3). This role for NMDARs is also specific for memory phase (LTM) and brain region (ellipsoid body). Initial learning and early memories were not affected when NMDARs were knocked down in the ellipsoid body, and LTM was not affected when NMDARs were knocked down in the mushroom body (Table 1). These results, together with recent observations that LTM formation may require neuronal activity from the vertical lobes of the mushroom body and correlates with the appearance of a Fas II–immunoreactive asymmetrical body near the central complex18,34,35, support a broader neuroanatomical circuitry involving both the mushroom body and ellipsoid body that subserves olfactory memory consolidation.
We also postulate that NMDARs in the ellipsoid body are involved with LTM consolidation and storage, but not retrieval. Disruption of NMDAR function during training, but not during testing, severely diminished LTM, excluding the possibility that memory retrieval was affected (Fig. 3c,d). Reversal memory was stronger when NMDARs were disrupted during an initial spaced training session compared with disruption during reversal training, directly demonstrating these flies’ ability to retrieve memory (Fig. 3e). That retrieval is normal and that consolidation and storage have failed is consistent with our initial study1 and with various studies of mammals36.
Blocking synaptic output from the mushroom body, but not from the ellipsoid body, during training and in the first 6 h after training abolished consolidation of LTM (Fig. 5b), suggesting that consolidation of LTM occurs downstream of the mushroom body (or possibly in the mushroom body if the efferents from the ellipsoid body were to project back to the mushroom body). In contrast, blocking synaptic output from the ellipsoid body or mushroom body disrupted retrieval, but not acquisition and consolidation (Fig. 5c). This result is functionally analogous to STM, which is formed in or upstream of the mushroom body, probably without the involvement of extrinsic neurons or structures30,32,37, and synaptic output from the mushroom body is required specifically for its retrieval, but not its acquisition38,39. Taken together, our observations support a model where memory is first acquired in the mushroom body and is then transferred to the ellipsoid body for storage during memory consolidation, in agreement with various observations from other species where memory transfer from one brain region to another may occurr6,33,36. Combined with other recent studies, we would propose (i) that acquisition involves an initial association of an odor and shock in and/or upstream of the mushroom body5,10, (ii) that STM resides in the mushroom body30,32,37, (iii) that MTM is acquired in mushroom body α/β neurons and persists there via recurrent activity involving the mushroom body α′/β′ DPM (dorsal-paired medial) neurons and the mushroom body α/β neurons themselves40, (iv) that LTM consolidation involves the transference of memory to the ellipsoid body and (v) that LTM retrieval requires neural activity output from the ellipsoid body and from mushroom body neurons34. Therefore, our results support a broader neuroanatomical circuitry involving both the mushroom body and ellipsoid body that subserves memory processing and retrieval.
Consolidation is the progressive stabilization of memory from a short, labile form to a long-lasting, stable form. Memory consolidation commonly refers to two types of processes, early and late consolidation. Early (or cellular or local) consolidation is accomplished within the first few minutes to hours after learning and occurs in all of the species studied to date41. This relatively fast type of consolidation takes place in local nodes in the neuronal circuit(s) and depends on cross talk among synapses, somata and nuclei41,42. Late consolidation, which so far has only been demonstrated in humans and mammals, takes much longer and involves multiple brain systems36,41–43. Late consolidation is initiated in parallel to, or as a consequence of, early consolidation, and is characterized by much slower temporal kinetics. Recent molecular and cellular studies in rodents have shown that a memory initially depends on the hippocampus, but eventually becomes independent of hippocampal function and may be consolidated into neocortical circuits36,41–43. To our knowledge, the existence of multiple memory systems has not clearly been demonstrated in any invertebrate model system. Our demonstration that NMDARs are specifically required in the ellipsoid body rather than in the mushroom body during LTM consolidation shows that memory consolidation is a systems-level phenomenon in Drosophila.
It has recently been shown that LTM requires branch-specific neural activity and CREB in α/β mushroom body neurons35. Notably, the only GAL4 driver used in that study, C739, is not specific to the mushroom body; it also labels some ellipsoid body neurons that appear to be different from the NMDAR-positive R4m neurons (Supplementary Figs. 5 and 9 online). Consistent with this observation, C739-targeted knockdown of functional NMDARs left LTM intact (Supplementary Fig. 8). When neural activity was silenced in UAS-shits1/+; C739/+ flies, however, memory retrieval was disrupted34. Considering that the cyclic AMP–dependent neuronal activity in the mushroom body is required for retrieval of LTM (S.X., H.W. and T.T., unpublished data), these results raise the interesting possibility that neural activity in the mushroom body α lobes may be correlated with memory retrieval rather than memory consolidation.
An intriguing difference exists in systems-level memory processing between Drosophila and rodents. Though LTM may eventually recruit the ellipsoid body, the mushroom body appears to be crucial for both memory consolidation and retrieval in Drosophila. In contrast, the hippocampus is required for consolidation, but not for retrieval, of long-lasting memories transferred to cortical systems36,41–43. This difference may highlight an unusual aspect of olfactory memory in Drosophila. We recently have found that the encoding of odor identity and odor intensity is experience-dependent and mushroom body–dependent44. Thus, the mushroom body appears to be central to olfactory discrimination as a perceptual task, providing a possible explanation as to why the mushroom body has to be involved with retrieval of LTM.
NMDARs in the mushroom body and MTM formation
The mushroom body is clearly involved in olfactory learning, memory consolidation and retrieval5,6,10. Nevertheless, consolidation of LTM occurred normally even when the function of NMDARs in the mushroom body was disrupted (Table 1). MTM depends on NMDAR function in the mushroom body (Fig. 6) and on amnesiac (amn)-encoded neuropeptides45 expressed in DPM neurons17. DPM neurons are extrinsic to the mushroom body, but nevertheless send extensive arborizations to all the mushroom body lobes17. Synaptic output from DPM neurons is required for the persistence of MTM, but does not appear to be involved in its acquisition and retrieval46. Consistently, MTM is proposed to be formed in the mushroom body α/β neurons, and blocking mushroom body output diminishes the retrieval of MTM34,40. Taken together, these observations suggest that the involvement of the mushroom body in LTM consolidation may be through an NMDAR-independent pathway, and that the mushroom body and ellipsoid body may participate independently in distinct temporal stages of memory consolidation, a neurobehavioral phenomena that has been reported in other species9,47.
Understanding how the NMDAR-containing protein complexes are independently involved, at the cellular level, with different memory phases in distinct brain regions, and how these distinct anatomical sites communicate with each other to yield adaptive behavior from prior experience will be of particular importance in the future.
METHODS
Fly stocks
Flies used were wild-type Canton-S w1118 (iso1CJ), the ‘Cantonized’ UAS-dsNR1, UAS-dsNR2, UAS-shits1, tub-GAL80ts, Feb170, Ruslan, C42, OK107, 201Y, C739, C155 (elav-GAL4), hs-GAL4 (P26) and UAS-mCD8::GFP. Feb170 and C42 were from G. Korge (Freie Universitaet Berlin), hs-GAL4 from Y. Zhong (Cold Spring Harbor Laboratory), UAS-mCD8::GFP from L. Luo (Stanford University) and tub-GAL80ts from Bloomington Fly Center. The genetic backgrounds for all stocks were Cantonized by outcrossing their heterozygous virgins with w1118 (iso1CJ) males for six generations.
Antibody production and western blotting
Rabbit antibodies to dNR1 have been described previously1 or are described in Supplementary Figs. 1 and 2. Rabbit polyclonal antibodies to dNR2 were raised against various peptides corresponding to different regions of dNR2 (Supplementary Figs. 1 and 2).
Western analysis was carried out as previously described1. In brief, adult heads were homogenized with lysis buffer, centrifuged at 14,000 rpm at 4 °C for 50 min, and the supernatant was saved. Lysate proteins were electrophoresed on a 6% SDS-PAGE gel and then electroblotted onto PVDF membranes. Immobilized proteins were probed with a rabbit polyclonal antibody to dNR1 (α-85S at 1:5,000 dilution) polyclonal antibodies to dNR2 (α-820-2 at 1:5,000 dilution, α-820-1 at 1:5,000 dilution or α-84S at 1:5,000 dilution) or a mouse monoclonal antibody to actin (1:2,500 dilution, Developmental Studies Hybridoma Bank) as a loading controls and the membrane was incubated with HRP-conjugated goat antibody to rabbit (1:10,000 dilution) or mouse (1:5,000 dilution) IgG. The positive signal was visualized with Qentix Western signal enhancer and SS West Pico Substrate detection (Pierce).
Behavior
Olfactory associative learning was measured by training 2–3-day-old adult flies in a T-maze with a Pavlovian conditioning procedure48. Groups of about 100 flies received one or ten sessions of massed or spaced training during which they were exposed sequentially to one odor (conditioned stimulus, CS+; 3-octanol or 4-methyl-cyclohexanol) paired with electric shock and then to a second odor (CS−; 4-methyl-cyclohexanol or 3-octanol) without eletric shock. Learning was measured immediately after one training session1. We tested 3-h memory after one training session8. We evaluated 1-d or 4-d memory after spaced or massed training, which induces strong, long-lasting memory for conditioned avoidance8. Spaced training consisted of ten cycles of one session of training with a 15-min rest interval between each cycle. Massed training consisted of ten cycles of one session of training with no rest interval between each. Afterwards, flies were tested for memory retention of conditioned avoidance at the choice point of the T-maze after 1 or 4 d. A performance index was calculated as the number of flies avoiding the CS+ minus that avoiding the CS−, divided by the total number of flies and finally mutiplied by 100.
Heat-shock regimen
Heat shock induction was carried out according to an established protocol with the same hs-GAL4 driver (P26)1.
Drug feeding
CXM feeding was carried out according to an established protocol8.
Statistics
As a result of the nature of their mathematical derivation, performance indexes are distributed normally. Hence, the data were evaluated via one- or two-way ANOVAs. Subsequent pair-wise planned comparisons were adjusted for experiment-wise error (α′), keeping the overall α = 0.05. All data were presented as mean ± s.e.m.
Methods used for the generation of flies carrying UAS-dsNR1 and UAS-dsNR2 transgenes, antibody production, immunohistochemistry and imaging whole-mount mCD8::GFP expression are described in the Supplementary Methods online.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Korge, Y. Zhong, L. Luo and Bloomington Fly Center for fly stocks. We also thank M. Heisenberg, H. Cline and J. Dubnau for comments and discussion. This work was supported by grants to T.T. from the US National Institutes of Health and Dart Neurosciences, LLC, and to A.-S.C. from the National Science Council, the Brain Research Center of the University System of Taiwan and the Technology Development Program of Ministry of Economy.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
AUTHOR CONTRIBUTIONS: C.-L.W. and S.X. conceived and designed the experiments. C.-L.W., S.X. and T.-F.F. carried out the experiments with technique support from H.W., D.L. and Y.-H.C. C.-L.W., S.X. and A.-S.C. analyzed the data. C.-L.W. and S.X. graphed the data. S.X. and T.T. wrote the paper.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Xia S, et al. NMDA receptors mediate olfactory learning andmemory in Drosophila. Curr. Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts AC, Glanzman DL. Learning in Aplysia: looking at synaptic plasticity from both sides. Trends Neurosci. 2003;26:662–670. doi: 10.1016/j.tins.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Tsien JZ. Linking Hebb’s coincidence-detection to memory formation. Curr. Opin. Neurobiol. 2000;10:266–273. doi: 10.1016/s0959-4388(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 4.Allweis C. The congruity of rat and chick multiphase memory-consolidation models. In: Andrew RJ, editor. Neural and Behavioral Plasticity. New York: Oxford University Press; 1991. pp. 370–393. [Google Scholar]
- 5.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu. Rev. Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 6.Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr. Biol. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogan JH, et al. Spaced training induces normal long-term memory in CREB mutant mice. Curr. Biol. 1997;7:1–11. doi: 10.1016/s0960-9822(06)00022-4. [DOI] [PubMed] [Google Scholar]
- 8.Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 9.Eichenbaum H, Cohen NJ. From Conditioning to Conscious Recollection: Memory Systems of the Brain 583. New York: Oxford University Press; 2001. [Google Scholar]
- 10.Gerber B, Tanimoto H, Heisenberg M. An engram found? Evaluating the evidence from fruit flies. Curr. Opin. Neurobiol. 2004;14:737–744. doi: 10.1016/j.conb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 12.Jefferis GS, Marin EC, Watts RJ, Luo L. Development of neuronal connectivity in Drosophila antennal lobes and mushroom bodies. Curr. Opin. Neurobiol. 2002;12:80–86. doi: 10.1016/s0959-4388(02)00293-3. [DOI] [PubMed] [Google Scholar]
- 13.Hanesch U, Fischback K-F, Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 1989;257:343–366. [Google Scholar]
- 14.Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J. Neurosci. 1993;13:1852–1861. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comas D, Petit F, Preat T. Drosophila long-term memory formation involves regulation of cathepsin activity. Nature. 2004;430:460–463. doi: 10.1038/nature02726. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 17.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 18.Pascual A, Huang KL, Neveu J, Preat T. Neuroanatomy: brain asymmetry and long-term memory. Nature. 2004;427:605–606. doi: 10.1038/427605a. [DOI] [PubMed] [Google Scholar]
- 19.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Carthew RW. Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 22.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 23.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 24.Flockhart I, et al. Fly RNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:D489–D494. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, et al. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- 26.Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Renn SC, et al. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J. Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- 29.Yin JC, et al. Induction of a dominant-negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 30.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 31.Tang YP, et al. Genetic enhancement of learning and memory in mice. Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- 32.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 33.Dash PK, Hebert AE, Runyan JD. A unified theory for systems and cellular memory consolidation. Brain Res. Brain Res. Rev. 2004;45:30–37. doi: 10.1016/j.brainresrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Isabel G, Pascual A, Preat T. Exclusive consolidated memory phases in Drosophila. Science. 2004;304:1024–1027. doi: 10.1126/science.1094932. [DOI] [PubMed] [Google Scholar]
- 35.Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 36.Nakazawa K, McHugh TJ, Wilson MA, Tonegawa S. NMDA receptors, place cells and hippocampal spatial memory. Nat. Rev. Neurosci. 2004;5:361–372. doi: 10.1038/nrn1385. [DOI] [PubMed] [Google Scholar]
- 37.Akalal DB, et al. Roles for Drosophila mushroom body neurons in olfactory learning and memory. Learn. Mem. 2006;13:659–668. doi: 10.1101/lm.221206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval, but not acquisition, of memory. Nature. 2001;411:476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- 39.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 40.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu. Rev. Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- 42.Frankland PW, Bontempi B. The organization of recent and remote memories. Nat. Rev. Neurosci. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- 43.Wiltgen BJ, Brown RA, Talton LE, Silva AJ. New circuits for old memories: the role of the neocortex in consolidation. Neuron. 2004;44:101–108. doi: 10.1016/j.neuron.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Xia S, Tully T. Segregation of odor identity and intensity during odor discrimination in Drosophila mushroom body. PLoS Biol. 2007;5:e264. doi: 10.1371/journal.pbio.0050264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 46.Keene AC, et al. Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron. 2004;44:521–533. doi: 10.1016/j.neuron.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Emptage NJ, Carew TJ. Long-term synaptic facilitation in the absence of short-term facilitation in Aplysia neurons. Science. 1993;262:253–256. doi: 10.1126/science.8211146. [DOI] [PubMed] [Google Scholar]
- 48.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J. Comp. Physiol. [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.