Figure 5. Curve fit of class 6 EPSPs and effect of two concentrations of MLA.
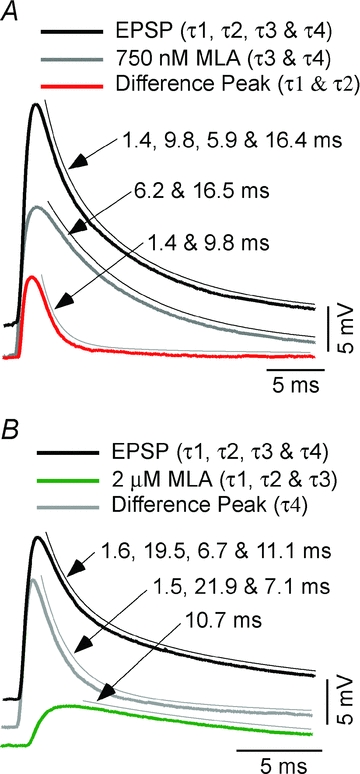
A, EPSP of class 6 CRN (thick black) is fitted as the sum of four decay exponentials τ1 = 1.4, τ2 = 9.8 ms, τ3 = 5.9 ms and τ4 = 16.4 ms with weights of 7.8, 1.0, 12.3 and 5.3 mV, respectively. The EPSP is partially blocked by 750 nm MLA, leaving a response (grey) fitted by two exponentials, τ3 = 6.2 and τ4 = 16.5 ms with weights of 11.7 and 5.0 mV, respectively. The difference trace (red), representing the component blocked by MLA, is fitted with a double exponential, τ1 = 1.4 and τ2 = 9.8 ms with relative weights of 7.9 and 1.0 mV. B, EPSP of class 6 CRN (thick black) is fitted as the sum of four decay exponentials τ1 = 1.6, τ2 = 19.5 ms, τ3 = 6.7 ms and τ4 = 11.1 ms with weights of 11.4, 2.6, 2.2 and 4.6 mV, respectively. The EPSP is partially blocked by 2 μm MLA, leaving an antagonist-insensitive component (green), fitted by a single exponential, τ4 = 10.7 ms with a weight of 4.5 mV. The difference trace (grey) is fitted by three exponentials, τ1 = 1.5, τ2 = 21.9 and τ3 = 7.1 ms with weights of 11.9, 2.3 and 2.1 mV, respectively, representing the components blocked by MLA. Stimulus and calibration artifacts are removed from the tail of each class 6 EPSP to facilitate curve fitting. All fits (thin black lines) are offset for clarity.
