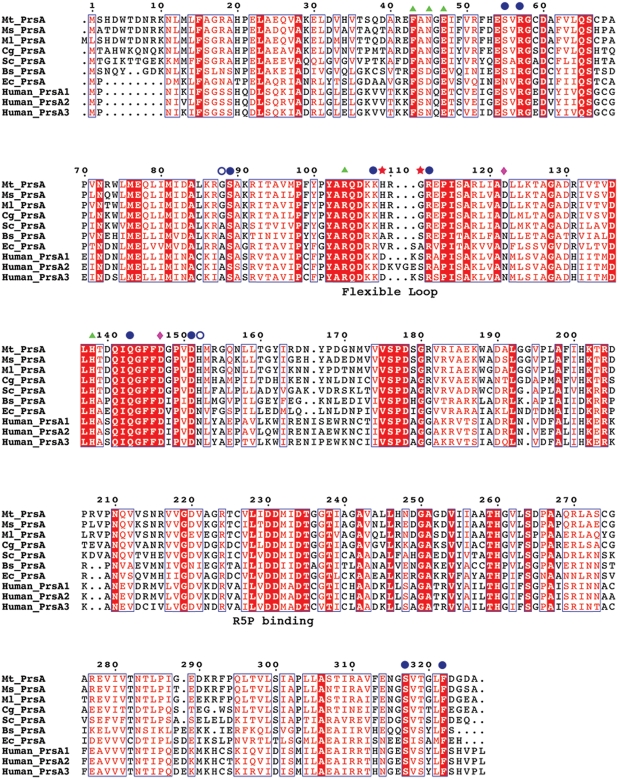
Fig. 10.
Sequence alignment of various prokaryotic pRpp synthetases. Protein sequences from M. tuberculosis (Mt_PrsA), M. smegmatis (Ms_PrsA), M. leprae (Ml_PrsA), C. glutamicum (Cg_PrsA), S. coleicolor (Sc_PrsA), B. subtilis (Bs_PrsA), E. coli (Ec-PrsA) and the three PrsA isoforms (Human_PrsA1–3) were aligned using CLUSTALW and annotated with ESPRIPT. Triangles represent residues involved in catalysis. Filled circles indicate conserved and open circles highlight nonconserved residues involved in allosteric regulation. His109 and Gly111 are highlighted with stars, indicating two residues only found in the flexible loop region of Corynebacterianeae PrsA homologs. The diamonds indentify the location of the residues involved in the formation of a salt bridge in Bs-PrsA and Human-PrsA1.