Histone lysine methylation (HKM) is a crucial epigenetic mechanism that establishes cell-specific gene expression and functions in development but is poorly understood in the retina. The authors examine the dynamic changes of specific HKM modifications and enzymes that control these marks in the developing retina and demonstrate a novel role for HKM in retinal neuron survival.
Abstract
Purpose.
Histone lysine methylation (HKM) is an important epigenetic mechanism that establishes cell-specific gene expression and functions in development. However, epigenetic control of retinal development is poorly understood. To study the roles of HKM in retinogenesis, the authors examined the dynamic changes of three HKM modifications and of two of their regulators, the histone methyltransferases (HMTases) Ezh2 and G9a, in the mouse retina.
Methods.
Retinal sections and lysates from embryonic day 16 through adult were processed for immunohistochemistry and immunoblotting using antibodies against various marks and HMTases. To further analyze the biological functions of HKM, the effects of small molecule inhibitors of HMTases were examined in vitro.
Results.
Methylation marks of trimethyl lysine 4 and 27 on histone H3 (H3K4me3 and H3K27me3) were detected primarily in differentiated retinal neurons in the embryonic and adult retina. In contrast, dimethyl lysine 9 on histone H3 (H3K9me2) was noted in early differentiating retinal ganglion cells but was lost after birth. The HMTases controlling H3K27me3, H3K9me2, Ezh2, and G9a were enriched in the inner embryonic retina during the period of active retinogenesis. Using the chemical inhibitors of Ezh2 and G9a, the authors reveal a role for HKM in regulating retinal neuron survival.
Conclusions.
HKM is a dynamic and spatiotemporally regulated process in the developing retina. Epigenetic regulation of gene transcription by Ezh2- and G9a-mediated HKM plays crucial roles in retinal neuron survival and may represent novel epigenetic targets to enhance viability in retinal neurodegenerative diseases such as glaucoma.
Genetic and epigenetic mechanisms ensure that complex developmental programs are correctly executed. Histone lysine methylation (HKM) is a crucial epigenetic mechanism that regulates gene transcription (activation and repression) and genome stability to influence development,1 stem cell pluripotency,2 tumorigenesis,3 and inflammation.4 Specific patterns of lysine methylation on histone H3 are associated with gene transcription and repression because of their effects on regulating the accessibility of particular DNA sequences, such as gene promoters and enhancers.5
HKM has been well studied at the K4, K9, and K27 residues. These lysine residues can be monomethylated, dimethylated, and trimethylated. Generally, trimethylation of lysine 4 on histone H3 (H3K4me3) is associated with fully activated promoters, which correlates with gene transcription, whereas dimethylation (H3K4me2) occurs at both inactive and active euchromatic genes.6,7 H3K9 is a major negative regulator of the H3K4 mark and dimethylation at lysine 9 (H3K9me2) marks silent euchromatin, important in proliferating cells, whereas H3K9me3 is enriched in regions of “gene-poor” pericentric heterochromatin.8–10 Methylation at lysine 27 on histone H3 (H3K27me) is associated with transcriptional repression in many developmental processes.11
The methylation of lysine residues on histone H3 is catalyzed by enzymes known as histone methyltransferases (HMTases). To allow for precise regulation, HMTases target specific lysine residues and methylation states. The activity of particular HMTases, such as G9a and Ezh2, which catalyze H3K9me2 and H3K27me3 marks,12,13 is essential to fundamental epigenetic processes such as X-chromosome inactivation14 and parental-specific silencing of imprinted genes, whose dysregulation is implicated in Prader-Willi syndrome.15 Moreover, pharmacologic inhibition of these HMTases is part of emerging therapeutic strategies to selectively inhibit tumor growth and reprogram easily isolable differentiated cells into induced pluripotent stem cells.16,17
The development of the retina is a highly regulated process whereby multipotent retinal progenitor cells generate a diverse, specialized set of glia and at least seven types of retinal neurons in a stereotypic temporal sequence.18 HKM is a key epigenetic mechanism regulating the ability of neural progenitor cells to self-renew and to generate neurons and glia in a precise temporal fashion.19,20 The lack of proper retinal cell maturation observed in zebrafish morpholinos, in which the function of the H3K9me3 HMTase Suv39h1 is disrupted, reveals a role for HKM in retinal development.21 Similar to the brain, most retinal neuronal types are born before the glial cells. For example, retinal ganglion cells (RGCs) are generated as early as embryonic day (E) 12, whereas Müller glial cell genesis peaks at postnatal day (P) 3.22 Other time-sensitive mechanisms operate. For instance, the ability of the newly born RGCs to robustly extend axons remains temporally limited. By E18, RGCs lose this intrinsic capacity.23 The retina has been long used to interrogate mechanisms underlying how common neural progenitors generate the cellular diversity and contribute to the function of a complex CNS structure.24 Interestingly, despite the facile accessibility of the retina and the increasing understanding of the role of epigenetics in regulating developmental competence, very little known about the retinal role of HKM and their master regulators, the HMTases.
We hypothesized that HKM is dynamically regulated in the developing retina. By immunostaining, we showed changing patterns of H3K4me3, H3K27me3, H3K9me2, Ezh2, and G9a in specific layers of the developing or adult retina. Interestingly, H3K9me2 marks were present in the inner embryonic retina but decreased rapidly by E18, a period during which RGCs lose the ability to extend axons. This mark nearly disappears, as does its corresponding HMTase, G9a, by adulthood. We also found that the master regulators of H3K27me3 and H3K9me2 marks, the HMTases Ezh2 and G9a, were enriched in the embryonic and neonatal period, during which retinal progenitor cells remained proliferative and neurogenic. Hypothesizing that these HMTases play important roles in the specific cell types that harbor their corresponding histone marks, we applied chemical inhibitors of Ezh217 and G9a25 to neonatal retinal neuronal cultures to reveal a new role for Ezh2 and G9a: RGC survival.
Materials and Methods
Primary RGC Isolation
All experimental procedures and use of animals were approved by the Animal Care and Use Committee of the Schepens Eye Research Institute and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. P0 mouse pups were anesthetized with hypothermia. The eyes were enucleated, and the retina was gently peeled off with fine forceps and placed in sterile phosphate-buffered saline (PBS). The retinas were collected in serum-free basal medium (Neurobasal A; Invitrogen, Carlsbad, CA) and incubated at 37°C with a papain dissociation system according to the manufacturer's instructions (Worthington, Lakewood, NJ). After 15 minutes of incubation, retinal digestion was stopped by the addition of the papain inhibitor ovomucoid (Worthington). RGCs were obtained by trituration of the retina in neuronal growth medium with a 1000-μL pipette. RGCs were isolated with magnetic bead-conjugated Thy1.2 antibody (Miltenyi Biotech, Auburn, CA) and were maintained in culture as described.26 Briefly, isolated RGCs were seeded in 24-well plates coated with poly-d-lysine (10 mg/mL; Sigma-Aldrich, St. Louis, MO) and cultured in serum-free basal medium (Neurobasal A; Invitrogen) supplemented with B27 and 100 U/mL penicillin-streptomycin (Invitrogen) as well as glutamine, brain-derived neurotrophic factor (50 ng/mL), ciliary neurotrophic factor (10 ng/mL), forskolin (5 ng/mL), and insulin (20 ng/mL) (all from Sigma-Aldrich). Cells were treated with BIX-01294 (10 nM, 50 nM, 100 nM, 200 nM) or DZNep (10 nM, 50 nM, 100 nM, 200 nM) for 48 hours.
RGC Apoptosis and Viability Analyses
Cellular apoptosis was determined using a fluorescein in situ cell death detection kit (Roche, Pleasanton, CA), which uses the incorporation of terminal transferase (TdT) to label free 3′OH ends in genomic DNA with fluorescein-dUTP (TUNEL) in apoptotic cells. To inhibit RGC apoptosis, 10 nM N-Benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone (Z-VAD-FMK or ZVAD; Sigma-Aldrich) was used.
Preparation of Retinal Sections
Retina sections were prepared as previously described.27,28 Briefly, the eyeballs from E16 to P0 were dissected, fixed in 4% paraformaldehyde for 1 hour, embedded in agarose, and sectioned at 100-μm thick using a vibratome. Adult mouse eyeballs were dissected, fixed in 4% paraformaldehyde for 1 hour, cryoprotected, embedded in optimal cutting temperature compound (Tissue Tek; Sakura Finetek, Torrance, CA), and cryosectioned at 8 μm.
Immunofluorescence Microscopy
For immunofluorescence labeling, retinal tissue sections or RGC cultures were blocked with blocking solution (5% bovine serum albumin + 5% normal goat serum + 0.3% triton X-100) for 1 hour at room temperature. The blocking buffer was discarded, and the sections were washed three times with 1× PBS, before the addition of antibodies (Millipore, Billerica, MA; Cell Signaling, Danvers, MA) against histone H3 lysine (K), methylation of trimethyl K4, dimethyl K9, and trimethyl K27 (all rabbit polyclonal, 1:200), Ezh2 (1:200, Cell Signaling; mouse monoclonal), and G9a (1:200; R&D Systems, Minneapolis, MN; mouse monoclonal). Retinal sections and cultures were also double labeled with primary antibodies against β-III-tubulin (Tuj1; 1:500; BD Biosciences, San Jose, CA), cellular retinaldehyde-binding protein (CRALBP), and rhodopsin (1:100; Chemicon, Temecula, CA). Incubation was performed overnight at 4°C. Sections were washed three times, followed by incubation with secondary antibody Cy-3 (1:200; Molecular Probes, Eugene, OR) conjugated with a fluorophore for 1 hour in the dark. The sections were washed again three times with 1× PBS for 30 minutes After staining with 4′,6-diamindino-2-phenyindole (2 mg/mL; Sigma) to reveal cell nuclei, retinal sections were mounted and examined under fluorescence and confocal laser scanning microscopy (SP5; Leica, Wetzlar, Germany). Quantification of fluorescent signal from anti-H3K27me3 and anti-H3K9me2 in RGCs in the presence and absence of Ezh2 inhibitor 3-Deazaneplanocin A (DZNep) and G9a inhibitor BIX-01294 was performed by determining the amount of fluorescence (in arbitrary units) using ImageJ software (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html) and dividing that value by the number of DAPI+ nuclei in the given field.
Preparation of Cell Lysates and Western Blot Analysis
Retinas from E16, E18, P0, and adult mice were dissected and homogenized in protein extraction reagent (B-PER; Pierce Biotech, Rockford, IL). Protein concentrations of retinal lysates were determined with a detergent-compatible colorimetric protein assay kit (DC Protein Assay Kit; Bio-Rad Laboratories, Hercules, CA). Proteins of retinal lysates (30 μg) were separated by an SDS-polyacrylamide gel and electrophoretically blotted onto a nitrocellulose membrane, incubated with mouse monoclonal Ezh2 antibody (1:500; Cell Signaling), and mouse monoclonal G9a antibody (1:500; R&D Systems). The blots were incubated with a horseradish peroxidase-conjugated second antibody (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA) and were detected by a chemiluminescence assay (ECL kit; Pierce). Histone H3 was used as the control for equal loading (1:500; Cell Signaling).
Statistical Analysis
In all experiments, mean ± SEM was presented as stated. Asterisks identify experimental groups significantly different from control groups by the Student's t-test. P < 0.05 was considered significant.
Results
Spatial and Temporal Regulation of HKM in the Retina
To identify patterns of HKM during retinal development, lysine methylation-specific antibodies were used to probe sections of embryonic (E16, E18), neonatal (P0), and adult (6-month-old) murine retinas. Ages for analysis encompass important development milestones, including RGC axonogenesis (E16 and earlier), RGC loss of axon growth capacity (E18), and photoreceptor genesis (perinatal and postnatal periods).22,29 Methylated H3K4, H3K9, and H3K27 marks are among the most well-studied HKM modifications in a variety of organ systems, animal and in vitro human models of development, and disease.11
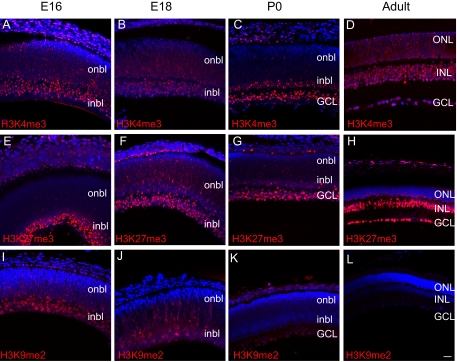
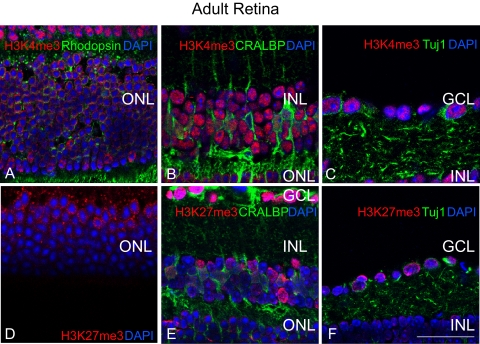
H3K4me3, a mark associated with active transcription,30 was present in RGCs (inner embryonic retina and ganglion cell layer [GCL] P0 and later) of the retina throughout the ages examined (Figs. 1A–D). In the E16 and E18 retina, the mark appeared to be enriched in the inner neuroblastic layer (inbl), where most postmitotic neurons reside (Figs. 1A, 1B). At P0, H3K4me3 was enriched throughout the GCL and the inbl; less expression of the mark was seen in other regions of the outer neuroblastic layer (onbl; Fig. 1C), similarly corresponding to regions of postmitotic neurons. In the adult retina, the H3K4me3 mark expanded to all layers of the neural retina (Fig. 1D), when the retina was largely composed of cells that exited the cell cycle. These included rhodopsin-positive photoreceptors (Fig. 2A), nearly all inner nuclear layer (INL) cells including CRALBP-positive Müller glia (Fig. 2B), and Tuj1-positive RGCs (Fig. 2C). In the adult, we observed that the H3K4me3 mark localized to the outer nuclear layer (ONL) periphery (Fig. 2A), whereas the mark in GCL and INL cells was distributed throughout the nucleus (Figs. 2B, 2C). These data show that H3K4me3, a euchromatic histone mark, is largely found in post-mitotic neurons in the inner and outer retinal layers throughout development and in the adult.
Figure 1.
HKM is spatially and temporally regulated in the developing and adult retina. (A–L) Immunofluorescence detection of H3K4me3 (A–D, red), H3K27me3 (E–H, red), and H3K9me2 (I–L, red) was overlaid on the nuclear counterstain (DAPI, blue) at E16, E18, P0, and adult. Scale bar, 20 μm.
Figure 2.
Adult retinal cells exhibit distinct nuclear patterns of HKM. (A–F) Euchromatic histone marks of activation (H3K4me3, red, A–C) and repression (H3K27me3, red, D–F) reveal distinct patterns of chromatin organization among adult retinal cell layers, including ONL photoreceptors (green, rhodopsin, A), INL Müller glia (green, CRALBP), and GCL RGCs (RGCs, green, Tuj1). Scale bar, 50 μm.
H3K27me3 is a mark associated with transcriptional repression, X-chromosome inactivation,14 body patterning,31 stem cell pluripotency,32 and other processes. To determine the spatial and temporal patterns of H3K27me3, a trimethyl-specific antibody for H3K27 was used to probe E16, E18, P0, and adult murine retinal sections. At E16 and E18, similar to that seen with H3K4me3, H3K27me3 marks were enriched in the inbl, though few scattered H3K27me3+ nuclei could be detected in the onbl (Figs. 1E, 1F). At P0, intensive labeling of the H3K27me3 mark was observed in the GCL and inbl (Fig. 1G). In the adult, GCL and INL nuclei strongly stained for the H3K27me3 mark (Fig. 1H). Further, a subset of nuclei in the inner portion of the ONL was H3K27me3+, and in these cells, similar to H3K4me3, the mark was also localized to the nuclear periphery (Fig. 2D), in contrast to GCL and INL nuclei (Figs. 1H, 2E 2F). These data show that the repressive H3K27me3 histone modification is enriched in the inner layers of the embryonic and adult retina and in a subset of ONL nuclei in the adult murine retina, which largely comprises a population of differentiated neurons.
H3K9me2 is a repressive mark known to silence euchromatic genes in embryonic cells.33 To examine for the presence of this histone modification, E16, E18, P0, and adult retinal sections were probed with a dimethyl-specific antibody against H3K9. Although the H3K9me2 mark was enriched in the E16 inbl, this mark appeared to decline abruptly at later time points (Figs. 1I–L). At E18, H3K9me2+ nuclei were still detectable in the inbl and onbl, though the intensity of staining significantly decreased (Fig. 1J). By P0, a few faintly stained H3K9me2+ nuclei were observed in the GCL, but this was extinguished in the adult retina (Figs. 1K, 1L). These results demonstrate that the repressive mark H3K9me2 is predominantly observed in the inner layers of the embryonic retina (inbl), but this modification declines in the late stages of retinogenesis. The spatiotemporal pattern of H3K9me2 is distinct from that of the H3K4me3 and H3K27me3 marks.
Detection of HMTases Ezh2 and G9a Expression in the Developing Retina
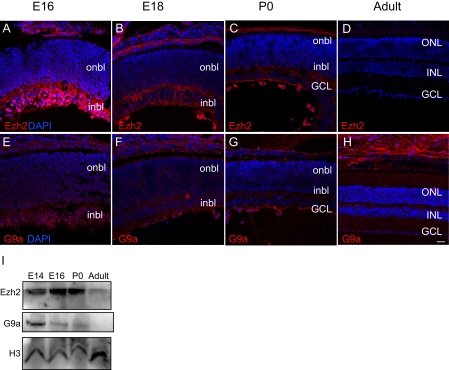
The HMTases constitute a family of enzymes that catalyze the methylation of histones on specific residues. The HMTases Ezh2 and G9a are the two best-characterized HMTases that catalyze H3K27me3 and H3K9me2 modifications, respectively. They are important in the repression of crucial genes in embryonic and tissue-specific development and homeostasis34,35 and regulate the differentiation of neural progenitors.36–38 To quantitatively examine their expression in the retina, we analyzed the temporal expression of Ezh2 and G9a by Western blotting whole retinal lysates isolated from mice at E14, E16, P0, and adult. Ezh2 was more highly expressed from E14 to P0, during the period of active retinogenesis, but its expression declined in the adult retina (Figs. 3A–D, 3I). To further characterize the spatial expression of this protein in the developing retina, we probed retinal sections with an antibody against Ezh2 (Figs. 3A–D) during the same time points used to analyze its downstream histone mark, H3K27me3 (Figs. 1E–H). Ezh2 was highly expressed in the inbl nuclei at E16, but the proportion of Ezh2+ nuclei declined rapidly at later time points, with little detectable expression in the adult retina. The high level of Ezh2 expression in the embryonic stages and its decreased presence in the adult suggests an active role for the protein during the period of retinogenesis.
Figure 3.
Ezh2 and G9a are preferentially expressed during retinogenesis. (A–H) Immunofluorescence detection of Ezh2 (A–D, red) and G9a (E–H, red) was overlaid on the nuclear counterstain (DAPI, blue) at E16, E18, P0, and adult. Scale bar, 20 μm. (I) Western blot analysis of Ezh2 and G9a at E14, E18, P0, and 6 months in whole retinal lysates.
Correlating with the temporal distribution of the H3K9me2 mark, the expression level of G9a—the HMTase responsible for H3K9me2—in whole retina lysates was higher during the period of retinogenesis (E14–P0) than in the adult (Figs. 3E–H, I). Specifically, G9a expression peaked at E14 and declined at later periods. Similarly, localization of the G9a in the developing retina showed the most prominent expression in the E16 inbl, but G9a+ nuclei were also observed in the E16 onbl (Fig. 3E). G9a+ nuclei rapidly decreased at E18 and P0, with little or no nuclear G9a seen in the adult retina (Figs. 3F–H). In contrast to H3K27me3 and Ezh2 (Figs. 1E–H, 3A–D), which demonstrated distinct spatiotemporal expression patterns from each other, the spatiotemporal pattern of G9a expression appeared similar to that of the H3K9me2 mark (Figs. 1I–L, 3E–H).
Regulation of RGC Survival by HMTases Ezh2 and G9a
A significant developmental event occurring in the mouse retina from E14 to P0 is the maturation of RGCs and the loss of axon growth capacity.29 Our results of immunohistochemistry and Western blot analysis indicated a close association with HKM, namely H3K9me2 and H3K27me3, and expression of the corresponding HMTases G9a and Ezh2, with RGCs during this period of retinal development. To interrogate the functional roles of HKM in retinal development, we sought to determine whether HMTases regulate RGC survival and differentiation. To this end, we isolated and cultured P0 murine RGCs26,39–41 in the presence or absence of small molecule inhibitors to G9a and Ezh2, BIX-01294 and 3-deazaneplanocin A, respectively.
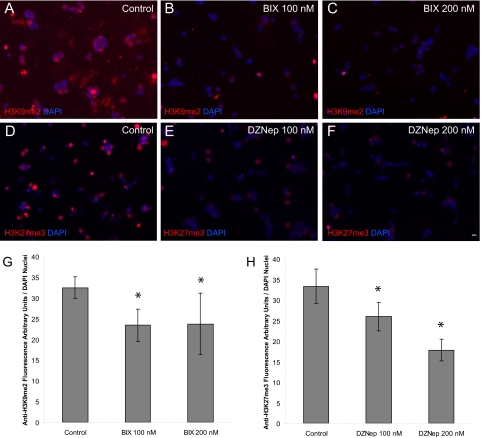
BIX-01294 is a diazepin-quinazolin-amine derivative that acts as a selective, reversible inhibitor of G9a and that has been shown to lower bulk H3K9me2 marks in several cell types.16,25,42 3-Deazaneplanocin A (DZNep) inhibits Ezh2-mediated H3K27 trimethylation.17,43 To determine whether BIX-01294 or DZNep lowers H3K9me2 or H3K27me3 levels, respectively, we cultured P0 RGCs in the presence and absence of these inhibitors and stained the cells with the corresponding marks. The amount of H3K9me2 fluorescence per nuclei decreased with BIX-01294 treatment (n = 2 per treatment group; P < 0.008; Figs. 4A–C, G), and the amount of H3K27me3 fluorescence per nuclei decreased with DZNep treatment (n = 2 per treatment group; P < 0.001; Figs. 4D–F, 4H).
Figure 4.
BIX-01294 (BIX) and DZNep inhibit H3K9me2 and H3K27me3 in P0 RGCs. (A–C) Immunofluorescence detection of H3K9me2 (red) and (D–F) H3K27me3 (red) with or without treatment of the described concentrations of BIX and DZNep was overlaid on the nuclear counterstain (DAPI, blue). Scale bar, 5 μm. (G) P0 RGCs were cultured for 3 days and treated with vehicle control, 100 nM and 200 nM BIX, or the same concentrations of (H) DZNep. Relative fluorescence per nuclei was obtained by dividing the fluorescence signal (arbitrary units) of the corresponding histone mark antibody by the total number of DAPI nuclei per 20× field using Image J software (mean ± SEM; n = 2 for each treatment group, eight fields per culture, mean 612 nuclei counted per field. *P < 0.008.
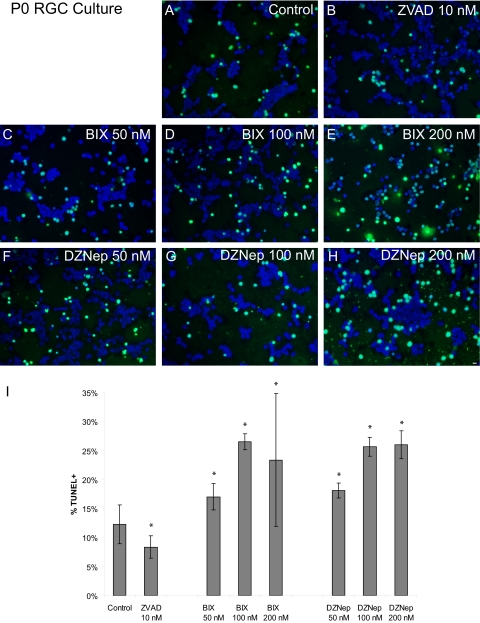
When RGCs were cultured in the presence of BIX-01294 and DZNep at 50 nM, 100 nM, and 200 nM, we detected RGC apoptosis. A roughly dose-dependent effect was observed compared with controls, with BIX-01294 inducing a 1.4- to 2.2-fold increase in apoptosis and DZNep inducing a 1.5- to 2.1-fold increase in apoptosis (n = 3 for each group; P < 0.015; Fig. 5). In contrast, the addition of the pan-caspase inhibitor (negative control) N-benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone (Z-VAD-FMK [ZVAD]) reduced apoptosis 1.5-fold compared with control cultures (Figs. 5B, 5I).
Figure 5.
Pharmacologic inhibition of Ezh2 and G9a induced RGC apoptosis. (A–H) P0 RGCs were cultured for 3 days with vehicle control, Z-VAD-FMK (ZVAD 10 nM), or increasing concentrations of BIX-01294 (BIX) and DZNep. TUNEL (green) and nuclei were counterstained with DAPI (blue). Scale bar, 5 μm. (I) Quantitation of (A–H). Apoptosis was analyzed by the TUNEL assay. Percentage of apoptotic cell death (mean ± SEM; n = 3 for each treatment group; 8 fields per culture, mean 740 nuclei counted per field; *P < 0.015) is expressed as the number of live cells divided by the total number of live and dead cells.
These results suggest an essential involvement of HKM in RGC survival. Inhibition of Ezh2 or G9a is associated with RGC death. These studies provide important information for future elucidation of the functional significance of these changes in retinal development and disease using mouse genetic technology.
Discussion
By monitoring HKM during various developmental stages, we explicitly defined patterns of H3K27me3, H3K9me2 marks, Ezh2, and G9a for the first time in the embryonic and adult retina and of H3K4me3 for the first time in the embryonic retina. We showed that these histone marks and HMTases were generally enriched in the inner layers of the embryonic retina; later, H3K4me3 and H3K27me3 modifications persisted in the adult neural retina. Interestingly, the H3K9me2 mark was largely lost in the adult retina. We also showed that the HMTases controlling H3K27me3, H3K9me2, Ezh2, and G9a were expressed in the embryonic and neonatal periods of retinogenesis, consistent with reports detailing these HMTases in other cell types and organ systems. Finally, through chemical inhibition of Ezh2 and G9a in cultures of neonatal RGCs, we showed that these HMTases are important for RGC survival.
We found that the patterns of H3K4me3 and H3K27me3, activating and repressive marks, overlapped in the embryonic and adult retina, especially in regions where postmitotic neurons exist. Of course, both these marks exist in most cells because they modify different regions of chromatin within a given cell. Future studies using chromatin immunoprecipitation (ChIP)-based technologies (e.g., ChIP-chip, ChIP-Seq) looking at specific, DNA-protein/histone modifications around key promoters required for retinal progenitor cell (Rx), RGC (Brn3b), INL neuron identity (amacrine/calbindin, horizontal/Prox1, bipolar/mGluR6, Müller /CRALBP), and photoreceptor (Crx) will further elucidate with greater specificity the epigenetic changes underlying the specification of diverse retinal cell types from progenitor cells and plasticity in postmitotic retinal cell types and mitotic adult glia such as Müller cells and retinal astrocytes.
A recent report44 described an association between nocturnal mammals (such as mice) and an inverted pattern of heterochromatin in adult rod nuclei.44 The sequestration of rod heterochromatin to a single, central chromocenter (highly stained, often “speckled” DAPI region, representing heterochromatin) is developmentally regulated and takes place approximately 1 month after birth. Our data confirmed that the H3K4me3 nuclear pattern is enriched in the euchromatic regions sequestered to the periphery of the ONL nuclei, surrounding the single murine rod chromocenter in the adult retina (Fig. 2A). Interestingly, we show that similar to H3K4me3, the repressive H3K27me3 mark is also enriched in the peripheral euchromatic regions of murine rod nuclei, consistent with the nuclear pattern of this mark observed in other tissues10 (Fig. 2D). The murine peripheral nuclear distribution of H3K4me3 and H3K27me3 appeared to be specifically restricted to ONL cells and developmentally regulated with their sequestration to the euchromatic ONL nuclear periphery not observed until sometime after P044 (Figs. 1C, 1D, 1G, 1H, 2A–F).
We noticed that G9a expression coincided with the H3K9me2 mark through time, namely a decreasing level of G9a and H3K9me2 as retinogenesis proceeded, with little or no G9a and H3K9me2 observed in the adult. The higher level of G9a at embryonic and neonatal stages was consistent with reports that G9a, more than other H3K9 HMTases such as Suv39h1/2, was especially critical during development in proliferating cells.8,45 How might the H3K9me2 retinal mark be lost through embryonic development? Possibilities include passive dilution of the modification after DNA replication in mitotic cells or through active enzymatic demethylation rather than decreased expression of the G9a HMTase.46–48 A similar mechanism may be responsible for the loss of the H3K9me2 mark in the retina, and it will be interesting to investigate whether H3K9me2 demethylases LSD1 or JHDM2A regulates the loss of the mark in this context.49,50
In the case of Ezh2 and H3K27me3, the level of Ezh2 decreased in the adult despite the persistence of the H3K27me3 mark in the GCL/RGC, INL, and some ONL nuclei. The persistence of the H3K27me3 mark in adult RGCs and inner retinal neurons may be secondary to the enhanced stability of the trimethyl (versus dimethyl or monomethyl) mark, retinal progenitor exit from the cell cycle (less DNA replication-associated dilution of the mark), or decreased activity of the H3K27me3 demethylases UTX and JHJD3.31,48,51–54 Additionally, it would be interesting to assess whether Ezh1 (an Ezh2 homologue) or H3K27me3 HMTase (largely expressed in adult nonproliferative cells) plays a role in regulating the H3K27me3 mark in the adult retina.55,56
We observed that the pharmacologic inhibition of G9a (through the addition of BIX-01294) in neonatal RGCs impairs RGC viability. These results are consistent with reports that genetic and chemical ablation of G9a deficiency results in somatic cell apoptosis in vivo and in vitro.8,25 Moreover, enhanced expression of the HMTase is involved in cell proliferation, as hypoxia induces G9a expression and increased H3K9me2, which is known to silence tumor suppressor RUNX3, and promotes tumor progression.57,58 Given the embryonic character of G9a expression in the inbl and its downregulation by E18, the period during which RGCs lose the ability to robustly extend their axons,59 its increased expression in proliferating cells,8 and the enhanced axonal regeneration of embryonic RGCs versus adult RGCs,60–62 it would be interesting to determine whether G9a overexpression stimulates RGC axonal regeneration.
The higher level of Ezh2 we observed during retinogenesis was consistent with the level found in previous reports demonstrating increased Ezh2 expression in embryonic and adult proliferating cells in other organ systems and tumors.35,37,56,63,64 Ezh2 is known to inhibit terminal differentiation in other organ systems such as the epidermal stem cell niche and in neural tumorigenesis.35,65,66 We found that Ezh2 was more highly expressed in immature, postmitotic RGCs in the E16 inbl than in more mature RGCs at later time points. It will be important to elucidate what function(s) Ezh2 may play in immature versus mature postmitotic RGCs. As in other regions of the CNS,36,37 the overexpression and genetic inhibition of Ezh2 in the retina may better clarify the role of this HMTase in regulating RGC development and retinal progenitor cell proliferation and the generation of RGCs and other retinal neurons from progenitor cells.
In conclusion, we describe for the first time embryonic (H3K4me3, H3K27me3, H3K9me2, Ezh2, G9a) and adult (H3K27me3, H3K9me2, Ezh2, G9a) HKM and HMTase patterns in the developing retina. We reveal a novel role for two important HMTases, G9a and Ezh2, in RGC survival. Further studies may implicate these histone marks and their HMTases in the epigenetic regulation of key cell-lineage genes during retinogenesis underlying retinal cell competence, adult retinal cell plasticity, retinal tumorigenesis (e.g., retinoblastoma), and retinal cell survival and axonal regeneration (e.g., retinitis pigmentosa and glaucoma).
Acknowledgments
The authors thank the members of the Chen Laboratory for critical discussion of the manuscript.
Footnotes
Supported by National Institutes of Health (NIH)/National Eye Institute (NEI) Grant R01EY017641, NIH/National Institute on Drug Abuse Grant R21DA024803, Department of Defense Grants W81XWH-04-2-0008 and W23RYX-9104-N603, Department of Veterans Affairs Grant 1I01RX000110-01, and the American Health Foundation (DFC); the Intramural Research Program of the NIH/National Cancer Institute Center for Cancer Research (VEM); NIH/NEI Grant P30EY003790; and a core grant to Schepens Eye Research Institute.
Disclosure: R.C. Rao, None; K.T. Tchedre, None; M.T.A. Malik, None; N. Coleman, None; Y. Fang, None; V.E. Marquez, None; D.F. Chen, None
References
- 1. Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135 [DOI] [PubMed] [Google Scholar]
- 2. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326 [DOI] [PubMed] [Google Scholar]
- 3. Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402 [DOI] [PubMed] [Google Scholar]
- 4. Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osipovich O, Milley R, Meade A, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5:309–316 [DOI] [PubMed] [Google Scholar]
- 6. Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are trimethylated at K4 of histone H3. Nature. 2002;419:407–411 [DOI] [PubMed] [Google Scholar]
- 7. Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766 [DOI] [PubMed] [Google Scholar]
- 8. Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rice JC, Briggs SD, Ueberheide B, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598 [DOI] [PubMed] [Google Scholar]
- 10. Peters AH, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589 [DOI] [PubMed] [Google Scholar]
- 11. Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124 [DOI] [PubMed] [Google Scholar]
- 12. Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317 [DOI] [PubMed] [Google Scholar]
- 13. Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J Biol Chem. 2003;278:14996–15000 [DOI] [PubMed] [Google Scholar]
- 16. Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574 [DOI] [PubMed] [Google Scholar]
- 17. Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46 [DOI] [PubMed] [Google Scholar]
- 19. Lim DA, Huang YC, Swigut T, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirabayashi Y, Suzki N, Tsuboi M, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613 [DOI] [PubMed] [Google Scholar]
- 21. Rai K, Nadauld LD, Chidester S, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205 [DOI] [PubMed] [Google Scholar]
- 23. Cho KS, Yang L, Lu B, et al. Re-establishing the regenerative potential of central nervous system axons in postnatal mice. J Cell Sci. 2005;118:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci U S A. 1987;84:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubicek S, O'Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481 [DOI] [PubMed] [Google Scholar]
- 26. Huang X, Wu DY, Chen G, Manji H, Chen DF. Support of retinal ganglion cell survival and axon regeneration by lithium through a Bcl-2-dependent mechanism. Invest Ophthalmol Vis Sci. 2003;44:347–354 [DOI] [PubMed] [Google Scholar]
- 27. Kinouchi R, Takeda M, Yang L, et al. Robust neural integration from retinal transplants in mice deficient in GFAP and vimentin. Nat Neurosci. 2003;6:863–868 [DOI] [PubMed] [Google Scholar]
- 28. Cho KS, Chen DF. Promoting optic nerve regeneration in adult mice with pharmaceutical approach. Neurochem Res. 2008;33:2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen DF, Schneider GE, Martinou JC, Tonegawa S. Bcl-2 promotes regeneration of severed axons in mammalian CNS. Nature. 1997;385:434–439 [DOI] [PubMed] [Google Scholar]
- 30. Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lan F, Bayliss PE, Rinn JL, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694 [DOI] [PubMed] [Google Scholar]
- 32. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194 [DOI] [PubMed] [Google Scholar]
- 35. Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–2883 [DOI] [PubMed] [Google Scholar]
- 38. Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem. 2009;284:2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu DY, Zheng JQ, McDonald MA, Chang B, Twiss JL. PKC isozymes in the enhanced regrowth of retinal neurites after optic nerve injury. Invest Ophthalmol Vis Sci. 2003;44:2783–2790 [DOI] [PubMed] [Google Scholar]
- 40. Kwong JM, Lalezary M, Nguyen JK, et al. Co-expression of heat shock transcription factors 1 and 2 in rat retinal ganglion cells. Neurosci Lett. 2006;405:191–195 [DOI] [PubMed] [Google Scholar]
- 41. Jiao J, Huang X, Feit-Leithman RA, et al. Bcl-2 enhances Ca(2+) signaling to support the intrinsic regenerative capacity of CNS axons. EMBO J. 2005;24:1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang Y, Zhang X, Horton JR, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glazer RI, Hartman KD, Knode MC, et al. 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem Biophys Res Commun. 1986;135:688–694 [DOI] [PubMed] [Google Scholar]
- 44. Solovei I, Kreysing M, Lanctot C, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368 [DOI] [PubMed] [Google Scholar]
- 45. Peters AH, O'Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337 [DOI] [PubMed] [Google Scholar]
- 46. Paik WK, DiMaria P. Enzymatic methylation and demethylation of protein-bound lysine residues. Methods Enzymol. 1984;106:274–287 [DOI] [PubMed] [Google Scholar]
- 47. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953 [DOI] [PubMed] [Google Scholar]
- 48. Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206 [DOI] [PubMed] [Google Scholar]
- 49. Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495 [DOI] [PubMed] [Google Scholar]
- 50. Perillo B, Ombra MN, Bertoni A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206 [DOI] [PubMed] [Google Scholar]
- 51. Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734 [DOI] [PubMed] [Google Scholar]
- 52. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094 [DOI] [PubMed] [Google Scholar]
- 53. Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450 [DOI] [PubMed] [Google Scholar]
- 54. Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300 [DOI] [PubMed] [Google Scholar]
- 55. Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee SH, Kim J, Kim WH, Lee YM. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–194 [DOI] [PubMed] [Google Scholar]
- 58. Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016 [DOI] [PubMed] [Google Scholar]
- 59. Chen DF, Tonegawa S. Why do mature CNS neurons of mammals fail to re-establish connections following injury—functions of bcl-2. Cell Death Differ. 1998;5:816–822 [DOI] [PubMed] [Google Scholar]
- 60. Vanselow J, Müller B, Thanos S. Regenerating axons from adult chick retinal ganglion cells recognize topographic cues from embryonic central targets. Vis Neurosci. 1991;6:569–576 [DOI] [PubMed] [Google Scholar]
- 61. Avwenagha O, Campbell G, Bird MM. The outgrowth response of the axons of developing and regenerating rat retinal ganglion cells in vitro to neurotrophin treatment. J Neurocytol. 2003;32:1055–1075 [DOI] [PubMed] [Google Scholar]
- 62. Park KK, Liu K, Hu Y, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Holling TM, Bergevoet MW, Wilson L, et al. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J Immunol. 2007;179:5317–5325 [DOI] [PubMed] [Google Scholar]
- 64. Chen H, Gu X, Su IH, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Richter GH, Plehm S, Fasan A, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]