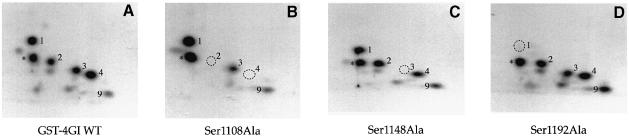
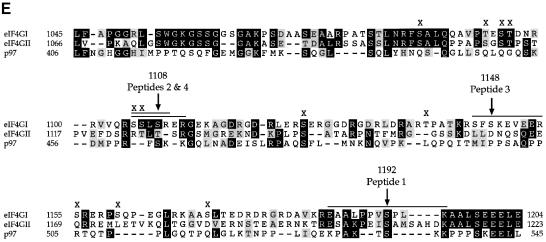
Fig. 5. Identification of serum-inducible phosphorylation sites by mutational analysis. Mutations of serine residues identified by mass spectrometry as phosphorylation sites were introduced into the eIF4GI phospho-region (aa 1035–1206)–GST fusion proteins. Proteins were expressed in 293T cells, 32P–labeled, isolated and mapped. Tryptic maps of the GST–eIF4GI (aa 1035–1206) wild-type protein (A), and point mutants Ser1108Ala (B), Ser1148Ala (C) and Ser1192Ala (D). A novel phosphopeptide that does not co-migrate with any endogenous eIF4GI peptide is indicated with an asterisk. (E) Complete mutational analysis summary. An alignment of the eIF4GI phospho-region with the corresponding regions of eIF4GII and p97. Identical residues are boxed in black, similar residues are boxed in gray. eIF4GI residues mutated to alanine are indicated by an ‘x’ or arrow. Locations of the serum-stimulated, phosphorylated residues, as well as the phosphopeptides identified in this analysis, are also indicated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.