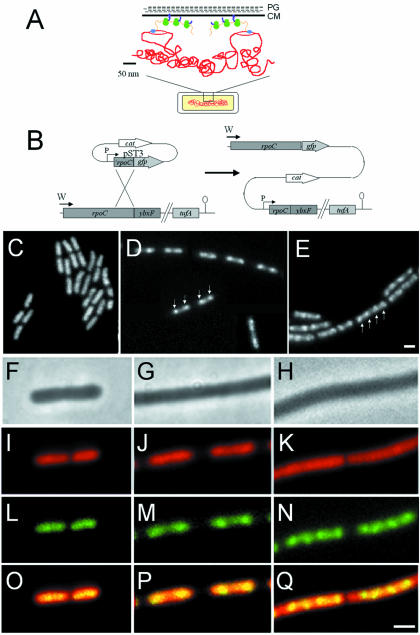
Fig. 1. (A) Schematic representation of transcription and translational organization in bacteria (derived from Woldringh et al., 1995). PG, peptidoglycan; CM, cytoplasmic membrane. Gram-positive organisms do not contain an outer membrane. The boxed region of the cell (bottom) is shown in expanded form above. Transcription by RNAP (blue) from DNA (red) that has looped out from the dense central mass of the nucleoid is closely linked to translation by ribosomes (green) via mRNA (orange). In this figure, the translated proteins (purple) are being inserted simultaneously into the cell membrane, but cytoplasmic proteins have also been proposed to be transcribed and translated in a similar peripheral subcellular location (Woldringh et al., 1995). (B) Construction of the fusion strain 1048 by insertion of pST3 into the chromosome. Integration of pST3 fuses the full-length rpoC gene, under the control of its natural promoter (W), to gfp. The plasmid provides a xylose-inducible promoter (P) to drive transcription of downstream genes, beginning with a truncated copy of rpoC. tufA is the last gene of the operon and encodes elongation factor Tu. (C–E) Images of RNAP–GFP in cells grown at 37°C in S medium [(C) doubling time 70 min], CH medium [(D) doubling time 45 min] and 2TY medium [(E) doubling time 20 min]. (D) is a composite image due to the low density of cells in the sample. (F–Q) Further magnified images of typical cells. (F), (I), (L) and (O) Cells grown in S medium. (G), (J), (M) and (P) Cells grown in CH medium. (H), (K), (N) and (Q) Cells grown in 2TY medium. (F–H) Phase contrast images; (I–K) DAPI images false coloured red; (L–N) RNAP–GFP images false coloured green; (O–Q) overlays of DAPI and GFP images. Scale bar, 2 μm.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.