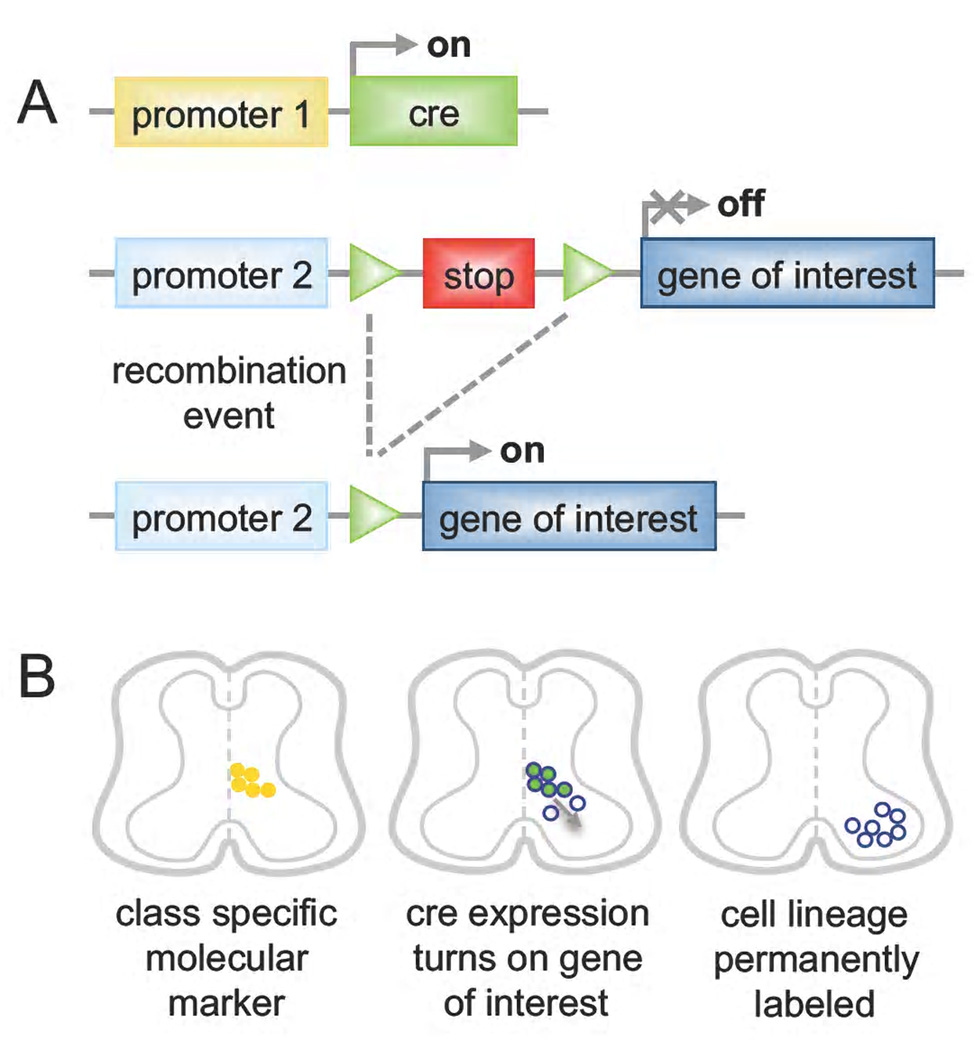
Figure 3. Cre-dependent manipulation of defined neuronal populations.
A) Two different alleles are used in combination to genetically manipulate defined populations of cells. First, the site-specific Cre recombinase is expressed from a promoter (1) specific to one neuronal class. A second allele is used to express a gene of interest from a promoter (2) that is, in general, constitutive and broadly active but can also be neuronal or cell type specific. Transcriptional STOP sequences flanked by loxP sites (green triangles) are interposed between the promoter and the coding sequence such that Cre-mediated recombination of loxP sites and excision of the STOP cassette is required to express the gene of interest. This strategy can be used to express a reporter gene such as GFP for morphological and axonal characterization, and fate mapping of the genetic lineage or to express an effector gene like TeNT or DTA, resulting in cell silencing or ablation for functional studies. B) Schematic picture showing genetic lineage labeling in the spinal cord. The promoter (1) of a molecular marker defining a neuronal class (yellow dots) is used to drive Cre expression in the same cells (green dots). Recombination induces expression of the gene of interest (blue circles), which is dependent on promoter (2). At later stages of development recombined cells have migrated to settle in their final location. Although these cells no longer express the molecular marker nor Cre, they are permanently labeled (blue circles).