Abstract
Rationale: Severe asthma (SA) remains poorly understood. Mast cells (MC) are implicated in asthma pathogenesis, but it remains unknown how their phenotype, location, and activation relate to asthma severity.
Objectives: To compare MC-related markers measured in bronchoscopically obtained samples with clinically relevant parameters between normal subjects and subjects with asthma to clarify their pathobiologic importance.
Methods: Endobronchial biopsies, epithelial brushings, and bronchoalveolar lavage were obtained from subjects with asthma and normal subjects from the Severe Asthma Research Program (N = 199). Tryptase, chymase, and carboxypeptidase A (CPA)3 were used to identify total MC (MCTot) and the MCTC subset (MCs positive for both tryptase and chymase) using immunostaining and quantitative real-time polymerase chain reaction. Lavage was analyzed for tryptase and prostaglandin D2 (PGD2) by ELISA.
Measurements and Main Results: Submucosal MCTot (tryptase-positive by immunostaining) numbers were highest in “mild asthma/no inhaled corticosteroid (ICS) therapy” subjects and decreased with greater asthma severity (P = 0.002). In contrast, MCTC (chymase-positive by immunostaining) were the predominant (MCTC/MCTot > 50%) MC phenotype in SA (overall P = 0.005). Epithelial MCTot were also highest in mild asthma/no ICS, but were not lower in SA. Instead, they persisted and were predominantly MCTC. Epithelial CPA3 and tryptase mRNA supported the immunostaining data (overall P = 0.008 and P = 0.02, respectively). Lavage PGD2 was higher in SA than in other steroid-treated groups (overall P = 0.02), whereas tryptase did not differentiate the groups. In statistical models, PGD2 and MCTC/MCTot predicted SA.
Conclusions: Severe asthma is associated with a predominance of MCTC in the airway submucosa and epithelium. Activation of those MCTC may contribute to the increases in PGD2 levels. The data suggest an altered and active MC population contributes to SA pathology.
Keywords: prostaglandin D2, chymase, carboxypeptidase A
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Severe asthma remains poorly understood. Mast cells (MCs) are implicated in asthma pathogenesis, but it remains unknown how their phenotype, location, and activation relate to asthma severity.
What This Study Adds to the Field
This study reports changes in phenotype profile, distribution, and activity of airway resident MC populations from a large group of subjects with asthma and normal control subjects. The MC population in severe asthma is dominated by the chymase-positive phenotype in submucosa and more uniquely in the epithelium. This is associated with increased bronchoalveolar lavage fluid prostaglandin D2 (PGD2) levels in severe asthma as compared to other steroid-treated subjects with asthma. A greater proportion of chymase-positive MCs and increased PGD2 were identified as important predictors of severe asthma. Targeting PGD2 signaling pathways could be an option for treatment of severe asthma.
Severe asthma represents about 10 to 15% of the asthma population and yet remains poorly understood and treated (1, 2). Although a plethora of studies suggest alterations in mast cells (MCs) may underlie its pathogenesis, their specific role is unclear (3–8).
MCs are evolutionarily ancient cells with complex function, believed to differentiate dependent on the tissue microenvironment in which they reside. In humans, ultrastructural characteristics and differential immunohistochemical staining (IHC) for the MC-specific proteases tryptase and chymase have identified MCT (tryptase-positive only) and MCTC (both tryptase- and chymase-positive) phenotypes. In addition to chymase, MCTC express cathepsin G and carboxypeptidase A3 (CPA3) (9–11). Normally, MCT predominate in the lung, whereas MCTC represent less than 20% of lung MCs. The distribution of MC phenotypes within a particular organ/tissue varies as well (3–8). In intestines of mice, the epithelial MC phenotype differs from that of cells in deeper mucosal regions and changes further during a parasitic infection (12). In humans, increases in nasal epithelial MCs in allergic rhinitis and after an allergen challenge have also been reported (13, 14). Although lower airway epithelial MCs have rarely been studied, expression of MC-specific mRNA in human airway epithelial cell brushings was reported to differentiate subjects with asthma from normal subjects (3, 15, 16).
Although general quantitative and phenotype changes in total MCs have been reported with conditions such as asthma and COPD, functional characteristics of IHC-identified MCTC remain poorly understood (3–8, 17). In vitro studies show that the two MC phenotypes functionally differ. Chymase inhibitors block IgE-mediated histamine release from skin MCs (predominantly MCTC) but not from “normal” lung MCs (predominantly MCT) (18). Similar phenotypic differences in response to C5a and compound 48/80 are also seen (19, 20). In addition, generation of prostaglandin (PG) D2 may be more pronounced in MCTC and not always associated with MC degranulation (21, 22). Finally, corticosteroid (CS) therapy in asthma may decrease MCT, but not MCTC (23).
Based on the cited studies, it was hypothesized that there would be a shift in MC phenotype with increasing asthma severity, such that an MCTC-like phenotype would predominate in the airway submucosa and epithelium of subjects with severe asthma as compared with subjects with milder asthma and normal subjects. This deviation in MC phenotype and distribution would associate with an activation profile more typical of an MCTC than an MCT. To investigate this hypothesis, endobronchial tissue samples collected across the National Heart, Lung, and Blood Institute's Severe Asthma Research Program (SARP) from a range of subjects with asthma and normal control subjects were evaluated for: (1) submucosal and epithelial distribution of MCTot and MCTC, (2) mRNA expression of their enzymes, and (3) bronchoalveolar lavage (BAL) fluid levels of tryptase and prostaglandin D2 (PGD2) as a measure of MC-specific activity. Statistical models were applied to determine the relative importance of MCs and their phenotypes and activation in predicting the clinical severity of asthma.
Some of the results have been previously reported in the form of abstracts (24, 25).
METHODS
Subjects
Males and females between 18 and 65 years of age who smoked less than five total pack-years and none in the last year were included in the study. After signing informed consent, subjects completed comprehensive questionnaires, underwent baseline and post-bronchodilator spirometry, allergen skin prick testing, blood draw, and bronchoscopy that included airway mucosa biopsy, epithelium brushing, and BAL. Subjects were recruited from seven SARP sites. SARP is a loose network of site-focused studies that contribute samples and data to a central database. However, the specific interests of the sites remain distinct. The SARP sites in this study include the Brigham and Women's Hospital, Cleveland Clinic, National Jewish Health/University of Pittsburgh (site moved from Denver to Pittsburgh in 2006), University of Virginia, University of Wisconsin, Wake Forest University, and Washington University.
Normal subjects (NC) were healthy atopic or nonatopic individuals with normal lung function and without a history of chronic respiratory conditions, including rhinitis requiring topical corticosteroid treatment. Subjects with asthma were evaluated and classified according to the ATS workshop criteria into subjects with severe or not severe asthma (26, 27). Subjects with severe asthma required high doses of inhaled corticosteroids (ICS) and/or use of oral corticosteroid (OCS) therapy for 50% or more of the previous year. In addition, subjects with severe asthma exhibited two or more of seven minor criteria as outlined in Reference 26. Subjects with not severe asthma were further grouped based on lung function and use of ICS. Those with a prebronchodilator FEV1 greater than 80% predicted and on nonsteroid therapy only to control their symptoms were termed “Mild asthma/no ICS.” The group termed “Mild asthma/+ICS” had an FEV1 greater than 80% predicted and were using low-moderate doses of ICS, whereas “Moderate asthma” included subjects with an FEV1 less than 80% predicted, all of whom were treated with low-moderate dose of ICS, with or without a second controller agent (leukotriene modifier or long-acting β-agonist) (27).
Questionnaires
Information collected through questionnaires included general demographics, medical and smoking history, and frequency of asthma symptoms (see online supplement for details). Subjects also reported the need for three or more steroid bursts, asthma-related urgent care visits, or hospitalizations. Reporting any one or more of those events in the 12 months preceding bronchoscopy identified the subject as having a recent exacerbation.
Pulmonary Function and Atopy Testing
Pulmonary function testing was performed according to American Thoracic Society guidelines and as previously described (27). The parameters measured included prebronchodilator FEV1 and FVC (as % predicted) and FEV1/FVC. Serum IgE was measured and atopy was assessed by skin prick testing to 14 common aeroallergens (see online supplement for details).
Bronchoscopy, BAL, and Sample Processing
These procedures were performed across sites according to previously published protocols and the SARP manual of procedures (4, 6, 27, 28). Endobronchial tissue for immunohistochemistry (IHC) was collected at all the SARP sites listed above, whereas tissue and epithelial cell samples for mRNA were collected only at the National Jewish Health and then the University of Pittsburgh site. Epithelial cells were obtained by brushing the proximal airways as previously described (28). The cells were placed in Trizol (Invitrogen, Carlsbad, CA) for extraction of mRNA. Tissue for IHC was fixed in 10% buffered formalin and embedded in paraffin, and tissue for mRNA was placed in Trizol.
BAL cells and fluid were separated by centrifugation at 400 × g. BAL fluid aliquots were stored at −80°C and cytospins processed as previously described (see online supplement for details).
Immunostaining and Tissue Morphometric Analysis
Tissue sections (5 μm) were immunostained at the Cleveland Clinic site using an automated system (see online supplement for details). Positively stained cells were counted in the area between the subepithelial basement membrane and smooth muscle and within an intact epithelial layer, at 400× magnification. Analyzed tissue areas were measured using Image-Pro software (MediaCybernetics, Washington, D.C.) and cell counts reported per square millimeter of submucosa or, for epithelial cell counts, per 10-mm length of subepithelial basement membrane.
Quantitative Real-Time Polymerase Chain Reaction
Tissue and epithelial cell expression of tryptase, chymase and carboxypeptidase A3 (CPA3) mRNA was determined by real-time quantitative polymerase chain reaction (PCR) as described previously (see online supplement) (28).
BAL Fluid Tryptase and PGD2 Measurements
Both mediators were measured using ELISA. BAL fluid tryptase levels were measured in Dr. L. B. Schwartz's laboratory (Virginia Commonwealth University, Richmond, VA) with an in-house total tryptase ELISA after concentrating the BAL fluid 15-fold by lyophilization as described (29). BAL fluid PGD2 was measured at Elisatech (Denver, CO) using commercially available ELISA components (Cayman Chemical, Ann Arbor, MI). The samples were first purified/concentrated using C18 Sep-Pak cartridges (Waters Corp, Milford, MA) and then derivatized per the ELISA manufacturer's instructions. Detection limits for those assays were 20 pg/ml for tryptase and 3 to 5 pg/ml for PGD2.
MC Phenotypes by IHC and Real-Time Quantitative PCR
MC phenotype in tissue samples was assessed by a standard IHC approach, detecting tryptase-positive and, on a consecutive section, chymase-positive cells. As double labeling was not performed, tryptase-positive MCs generally reflect the total number of MCs (MCTot), including MCs that are tryptase-only positive (MCT; their low chymase levels are undetectable by routine IHC) and those that are both tryptase and chymase positive (MCTC) by routine IHC. Chymase-positive MCs in this study reflected the MCTC population. Consequently, MCTC/MCTot estimated the proportion of MCTC in the total MC population.
In addition to IHC, the MC presence was estimated by mRNA levels as measured by quantitative real-time PCR for the phenotype-identifying MC enzymes tryptase and CPA3. Chymase mRNA levels across all groups and compartments were very low. Therefore, the MCTC phenotype was estimated at the mRNA level through expression of CPA3 mRNA, another enzyme reported to differentiate the MCTC from a MCT phenotype (11). A small substudy (n = 14), which showed a significant correlation of chymase-positive MCs with CPA3-positive MCs in tissue by IHC (rs = 0.61; P = 0.02), supported such an approach.
Final Selection of Study Population
For reasons of the diverse interests of the SARP sites, not all subjects in the bronchoscopy data set (large SARP cohort with IHC and BAL data) had measurements available for all four MC markers (IHC tryptase and chymase, BAL fluid tryptase, and PGD2) analyzed in this report. Therefore, an a priori decision was made to only include subjects with data for at least two of the four MC markers (n = 157). The five groups were not statistically different in fractions of missing data for submucosal tryptase or chymase (overall P = 0.11 and 0.06, respectively), or for epithelial tryptase or chymase (overall P = 0.11 and 0.69, respectively). Details and Figure E1 showing total study population are in the online supplement.
Messenger RNA analysis was performed in subjects from the Pittsburgh site only. Due to the limited number of subjects with moderate asthma (n = 1) with tissue and/or epithelial mRNA, analysis was performed on NC, mild asthma/no ICS, mild asthma/+ICS, and severe asthma subjects (n = 60). Thirty-one subjects had mRNA for MC markers measured in both types of samples. As 18 subjects in the Pittsburgh mRNA cohort were also included in the large SARP cohort (they also had two or more IHC and/or BAL markers measured), a total of 199 subjects were included (see online supplement).
Statistical Analysis
Nonparametric Kruskal-Wallis tests were used to compare medians for the MC markers across the five groups. Analysis of variance was used to compare the means of the log-transformed mRNA data. When an overall significant difference was detected (P < 0.05), individual asthma groups were compared with NC (four comparisons) and the severe asthma group to each of the less severe groups (three comparisons). The Bonferroni procedure was used to conservatively test for the multiple post hoc comparisons between groups using an α = 0.05/7 = 0.0071 (for mRNA data, α = 0.05/5 = 0.01) for each comparison. To determine whether CS use impacted submucosal MC counts, MCTot were compared among the four asthma groups treated with increasing doses of CS, whereas MCTot and MCTC were compared in severe asthma treated or not with OCS. Categorical variables were compared by Pearson chi-square tests. Spearman correlations (rs) measured the association between any two MC markers and between MC markers and continuous measures of lung function among the whole population and, when significant, among the phenotypic groups. The small size of the moderate asthma group (n = 6–12) prevented subgroup analysis on that group. Normally distributed data (log-transformed mRNA data) were correlated using Pearson correlation coefficients (r).
General linear models and contrast coefficients were used to conduct tests for trend on MC markers that could be transformed to more normal distributions. The square root transformation was applied for MCTot, MCTC, and MCTC/MCTot and the natural logarithm transformation for tryptase and CPA3 mRNA levels. The IHC epithelial and BAL fluid markers showed a high proportion of the lowest detectable limit and high positive skew in their distributions. For these markers, the nonparametric Jonckheere-Terpstra test (JT) was used for testing trends across the five groups.
A multiple logistic regression model was used to determine the significance of the associations between the MC markers and severity of asthma (severe versus not severe) among the participants with asthma. Due to the amount of missing data, epithelial MC counts or any mRNA data could not be included in the analysis. Univariate analyses were first conducted for FEV1% predicted, maximum bronchodilator reversibility, BAL eosinophil percentage, the submucosal markers, BAL fluid PGD2, therapy with leukotriene modifiers, and atopy. Any variable significant at P less than 0.20 was entered into a multiple logistic regression model controlling for race and age. We controlled for age and race because both variables were important potential confounders as they were associated with asthma severity and several of the lung function and MC markers. Due to the high proportion of missing values for submucosal MCTC/MCTot, a sensitivity analysis was conducted using multiple imputation for the final logistic regression model. Results for the models are presented with odds ratios and 95% confidence intervals. The data were analyzed using SAS/STAT software, Version 9.1.3 of the SAS System for Windows.
RESULTS
Demographics
Participants were predominantly women (2:1) across all groups (Table 1). Subjects in the more severe groups were older compared with NC and mild asthma/no ICS. The groups differed by race, with a higher proportion of African Americans in the moderate and severe asthma groups. Atopy was less common in NC, but was similarly present among the asthma groups. Five subjects in the severe asthma group were on anti-IgE therapy (data not shown). Compared with the large SARP cohort, the University of Pittsburgh mRNA cohort's 9 subjects with mild/no ICS asthma were older, whereas the 23 subjects with severe asthma had a lower FEV1% predicted (median [25th–75th percentile] 49% [35–74] vs. 67% [56–82]; P = 0.005; Table 2). Systemic CS therapy was more common in the Pittsburgh severe asthma group (P = 0.02). The contribution of subjects from each of the participating centers is presented in Table 1.
TABLE 1.
SUBJECT CHARACTERISTIC OF THE LARGE SEVERE ASTHMA RESEARCH PROGRAM COHORT (N = 157)
| Normal Control Subjects (n = 34) | Mild Asthma No ICS (n = 22) | Mild Asthma on ICS (n = 31) | Moderate Asthma (n = 13) | Severe Asthma (n = 57) | Overall Difference (P Value) | |
|---|---|---|---|---|---|---|
| Age, yr* | 26 (24–34) | 23 (20–30) | 37 (23–40) | 38 (29–41) | 44 (32–50) | <0.0001 |
| Sex, % female | 62 | 59 | 65 | 77 | 68 | NS |
| AA/white/other, % | 12/70/18 | 14/77/9 | 16/81/3 | 46/54/0 | 37/58/5 | 0.01 |
| Atopy, % | 32.4 | 91 | 77.4 | 100 | 77 | <0.0001 |
| Serum IgE, kU/l* | 23 (13–53) | 80 (30–167) | 202 (70–352) | 150 (88–388) | 170 (62–344) | <0.0001 |
| Baseline FEV1% predicted* | 99 (93–107) | 97 (89–106) | 91 (84–104) | 69 (64–73) | 67 (57–83) | <0.0001 |
| Use of systemic CS therapy, n | N/A | 1 | 1 | 0 | 32 | <0.0001 |
| Use of LABA, % | N/A | 0 | 81 | 77 | 89 | <0.0001 |
| Use of LTM, % | N/A | 10 | 17 | 31 | 48 | 0.003 |
| Subjects by site, n (%) of each severity group | ||||||
| BWH | 0 | 0 | 6 (19) | 1 (8) | 1 (2) | |
| CLC | 8 (23) | 3 (14) | 1 (3) | 2 (15) | 6 (11) | |
| PITT | 15 (44) | 4 (18) | 1 (3) | 0 | 20 (35) | |
| UVA | 0 | 0 | 0 | 0 | 1 (2) | |
| WIS | 7 (21) | 8 (36) | 17 (55) | 3 (23) | 10 (17) | |
| WFU | 0 | 5 (23) | 4 (13) | 5 (38) | 6 (10) | |
| WSL | 4 (12) | 2 (9) | 2 (6) | 2 (15) | 13 (23) | |
Definition of abbreviations: AA = African American; BWH = Brigham and Women's Hospital Boston, MA; CLC = Cleveland Clinic, Cleveland, OH; CS = corticosteroids; F = female; ICS = inhaled corticosteroids; LABA = long-acting β-agonists; LTM = leukotriene modifiers; PITT = National Jewish Health, Denver, CO, and University of Pittsburgh, Pittsburgh, PA; UVA = University of Virginia, Charlottesville, VA; WIS = University of Wisconsin, Madison, WI; WFU = Wake Forest University, Winston-Salem, NC; WSL = Washington University, St. Louis, MO.
If not indicated otherwise, analyses were done using Pearson chi-square tests.
Data analyzed using Kruskal/Wallis tests and presented as medians (25th–75th percentile).
TABLE 2.
SUBJECT CHARACTERISTICS OF THE UNIVERSITY OF PITTSBURGH COHORT (N = 60)
| Normal Control Subjects (n = 21) | Mild Asthma No ICS (n = 9) | Mild Asthma on ICS (n = 7) | Severe Asthma (n = 23) | Overall Difference (P Value) | |
|---|---|---|---|---|---|
| Age, yr* | 29 (25–49) | 38 (27–57)† | 27 (21–31) | 39 (30–47) | 0.05 |
| Sex, % female | 43 | 78 | 71 | 61 | NS |
| AA/white/other, % | 5/71/24 | 33/56/11 | 14/72/14 | 26/74/0 | NS |
| Atopy, % | 52 | 100 | 85.7 | 74 | 0.05 |
| Serum IgE, kU/l* | 25 (14–42) | 137 (57–402) | 137 (47–341) | 154 (43–432) | 0.0004 |
| Baseline FEV1% predicted* | 100 (94–106) | 90 (82–96) | 103 (94–107) | 49 (35–74)† | <0.0001 |
| Use of systemic CS therapy, n | N/A | 0 | 0 | 17‡ | <0.0001 |
| Use of LABA, % | N/A | 14 | 71 | 87 | 0.0005 |
| Use of LTM, % | N/A | 14 | 14 | 65 | 0.007 |
Definition of abbreviations: AA = African American; CS = corticosteroids; F = female; ICS = inhaled corticosteroids; LABA = long-acting β-agonists; LTM = leukotriene modifiers; SARP = Severe Asthma Research Program.
If not indicated otherwise, analyses were done using Pearson chi-square tests.
Data analyzed using Kruskal/Wallis tests and presented as medians (25th–75th percentile).
P < 0.01 when compared to the SARP cohort.
P = 0.02 when compared to the SARP cohort.
Submucosal Tissue MCs: Severe Asthma is Associated with Reduced MCTot Numbers, but a Greater Proportion of MCTC
Immunohistochemistry for MCTot and MCTC in airway submucosa.
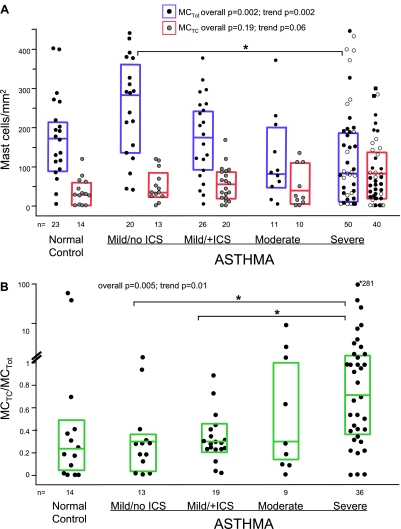
MCTot numbers in the proximal airway submucosa differed across the five subject groups (overall P = 0.002; Figure 1A). Submucosal MCTot were most numerous in mild asthma/no ICS (median [25th–75th percentile] 285/mm2 [135–361]) and were lowest in the severe asthma group (79/mm2 [9–188]). There was a significant trend for MCTot to lower levels with increasing asthma severity across all groups (Ptrend = 0.002).
Figure 1.
(A) Distribution of submucosal total mast cells (MCs) (tryptase-positive MCTot; solid circles/blue lines show median and 25th–75th percentiles) and chymase-positive MCs (MCTC; shaded circles/red lines show median and 25th–75th percentiles) evaluated by immunostaining. Open circles in the severe asthma group represent subjects who are not on systemic corticosteroid therapy. (B) Increase in MCTC/MCTot with increasing asthma severity. The groups were compared using Kruskal/Wallis tests (overall P values) and Jonckheere-Terpstra tests (trend P values). Marked are intergroup differences that remained significant after Bonferroni correction (P ≤ 0.007). ICS = inhaled corticosteroid.
Although MCTC counts did not differ between the groups (overall P = 0.19), there was a marginal trend for higher MCTC with increasing asthma severity (Ptrend = 0.06; Figure 1A). The overall reduction in MCTot and the concomitant development of a predominance of the MCTC phenotype with greater asthma severity resulted in a higher MCTC/MCTot in severe asthma (overall P = 0.005, Ptrend = 0.01; Figure 1B) than in mild asthma/no ICS (P = 0.004) or mild asthma/+ICS (P = 0.001).
Submucosal MCTot and MCTC were modestly correlated across all subject groups (rs = 0.34; P < 0.001). However, this positive association was only present in mild asthma/+ICS (rs = 0.78; P < 0.0001) and severe asthma (rs = 0.38; P = 0.02), suggesting that in these more severe CS-treated groups, a consistent shift in MCTot population toward MCTC phenotype occurs. Table E2.A presents correlations between IHC MC markers in different airway compartments.
Subgroup analysis for tissue mRNA.
Tissue biopsy samples for mRNA analyses were available from 34 subjects. Tissue tryptase and CPA3 mRNA levels did not differentiate NC (n = 14) from mild asthma/no ICS (n = 5), mild asthma/+ICS (n = 4), or severe asthma (n = 11) subjects (overall P = 0.35 and P = 0.36, respectively; data not shown). In contrast to IHC, tissue tryptase and CPA3 mRNA levels were strongly correlated (r = 0.81; P < 0.0001), supporting their common MC origin (Table E2.B). However, there were no significant relationships between submucosal MC numbers (by IHC) and levels of MC-specific markers (by mRNA).
Effect of CS therapy on MC numbers.
As CS therapy has been reported to impact submucosal MC counts, MC numbers were compared between the mild asthma groups on and off ICS, between subjects on low-dose ICS (mild) versus those on higher doses (moderate and severe), as well as between those subjects with severe asthma on and off OCS (23). The mild asthma/+ICS group, which did not differ in FEV1 compared with the mild asthma/no ICS, had significantly lower submucosal MCTot (P = 0.008) supporting an effect of ICS. However, the use of higher doses of ICS (and systemic CS) was not associated with lower MCTot numbers in moderate and severe asthma as compared with mild asthma/+ICS (overall P = 0.34). Finally, there were no differences in MCTot or MCTC numbers in subjects with severe asthma treated or not with systemic CS (P values both > 0.8).
Epithelial MCs: Severe Asthma is Associated with Higher Numbers of Epithelial MCTC Compared with Milder Asthma
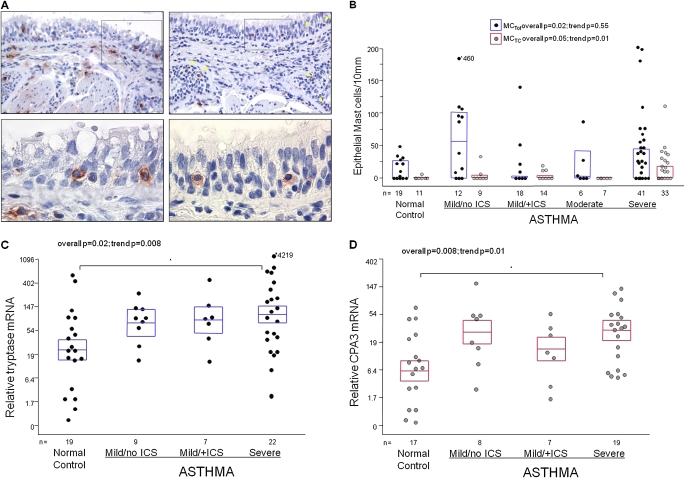
Epithelial MCTot varied with asthma severity (overall P = 0.02; Figures 2A and 2B). Similar to submucosal MCTot, epithelial MCTot were highest in mild asthma/no ICS (60/10 mm [2–105]). However, there was no trend for epithelial MCTot counts across the groups (JT test for trend P = 0.55). Submucosal and epithelial MCTot counts were moderately well correlated (rs = 0.49; P < 0.0001; Table E2.A). Epithelial MCTot and submucosal MCTot correlations were consistently high in the presence of CS treatment (mild asthma/+ICS rs = 0.53; P = 0.02, and severe asthma rs = 0.65; P < 0.0001). In contrast, the correlations were weak in NC or mild asthma/no ICS (rs all < 0.2).
Figure 2.
(A) Immunostaining for tryptase (left panels) and chymase (right panels) in the same proximal airway region from a subject with severe asthma. Pictures in upper panels are taken at 200× magnification. Arrows (yellow) in the right upper panel point at chymase-positive epithelial mast cells. Epithelial mast cells within boxes are shown in lower panels at 600× magnification. (B) Distribution of epithelial total mast cells (MCs) (tryptase-positive MCTot; black circles/blue lines are median and 25th–75th percentiles) and chymase-positive mast cells (MCTC; gray circles/red lines are medians and 25th–75th percentiles) evaluated by immunostaining. The groups were compared using Kruskal/Wallis tests (overall P value) and Jonckheere-Terpstra tests (trend P values). (C) Relative mRNA levels for tryptase, and (D) carboxypeptidase A3 (CPA3) in airway epithelial brushings. Y-axis represents a natural log-scale of the original linear values, with the means and SEM transformed from the natural log to the linear values. The groups in C and D were compared using analysis of variance and linear trend analysis. Marked are intergroup differences that remained significant after Bonferroni correction (P ≤ 0.01). ICS = inhaled corticosteroid.
Epithelial MCTC also differed across groups (P = 0.05; Figures 2A and 2B). Unlike MCTot, there was a significant trend to higher numbers from NC to severe asthma (JT test for trend P = 0.01). Epithelial MCTC were rarely detectable in NC (9% of subjects) but more likely to be seen in milder asthma (∼ 20% of subjects) and severe asthma (42% of subjects). The number of epithelial and submucosal MCTC marginally correlated overall (rs = 0.30; P = 0.009), without difference among the individual groups.
Epithelial Cell mRNA subgroup analysis.
Epithelial brushings were available from 19 NC, 9 mild asthma/no ICS, 7 mild asthma/+ICS and 22 severe asthma subjects. Epithelial tryptase and CPA3 mRNA increased in asthma and were highest in severe asthma (overall P = 0.02 and P = 0.008, respectively; Ptrend = 0.008 and Ptrend = 0.01, respectively; Figures 2C and 2D). The higher levels of mRNA for both markers in severe asthma paralleled the higher numbers of epithelial MCTC seen in the larger IHC database.
Epithelial tryptase mRNA consistently correlated with CPA3 mRNA (r = 0.74; P < 0.0001) across all subjects and in each individual group (r = 0.65–0.98). Despite being measured in different samples (biopsy tissue vs. brushings), tryptase and CPA3 mRNA correlated between epithelium and tissue (r = 0.73; P < 0.0001 and r = 0.57; P = 0.001, respectively; Table E2.B). There were no statistically significant correlations between MC counts (by IHC) in submucosa and epithelium (n = 18) and matching mRNA levels.
Activity of MCs: Severe Asthma Is Associated with Evidence for MCTC Activation
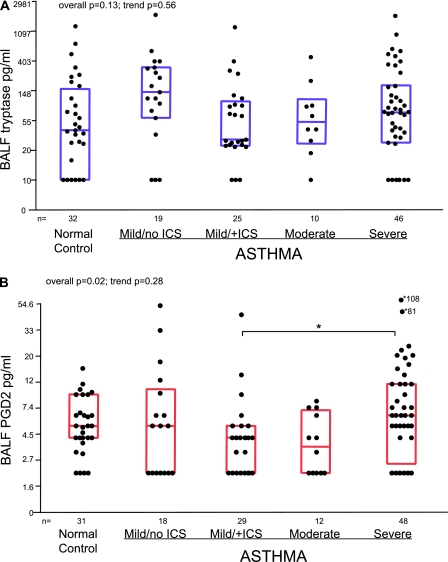
MC activity was assessed by comparing BAL fluid levels of tryptase (n = 132) and PGD2 (n = 138). BAL fluid tryptase did not differ among the five groups (overall P = 0.13; Figure 3A). Similar to submucosal and epithelial MCTot, BAL fluid tryptase was numerically highest in mild asthma/no ICS and there was no evidence for higher levels with increasing severity (JT test for trend P = 0.56).
Figure 3.
Concentrations of (A) tryptase and (B) prostaglandin D2 (PGD2) measured in bronchoalveolar lavage fluid (BALF) samples. Tryptase levels that were below the limit of detection were assigned a value of 10 pg/ml. PGD2 values that were below detection limit were given value of 2 pg/ml. Y-axis represents a natural log scale of the original linear values. The median and 25th–75th percentiles are transformed from the natural log to the linear values. The groups were compared using Kruskal/Wallis tests (overall P value) and Jonckheere-Terpstra tests (trend P values). Marked are intergroup differences that remained significant after Bonferroni correction (P ≤ 0.007). ICS = inhaled corticosteroid.
In contrast to tryptase, BAL fluid PGD2 differed significantly among the five groups (overall P = 0.02; Figure 3B). Paralleling the increase in submucosal and epithelial MCTC, PGD2 was higher in severe asthma as compared with other steroid-treated groups (6.0 pg/ml [2.5–11.0 pg/ml] vs. 4.0 pg/ml [2.0–6.0 pg/ml] in mild/moderate groups combined and 5.0 pg/ml [4.0–9.0 pg/ml] in normal controls, respectively; JT test for trend P = 0.28).
Although only BAL fluid PGD2 significantly differed between the groups, BAL fluid PGD2 and tryptase levels were modestly correlated (rs = 0.33; P < 0.001), suggesting a common MC source. Neither mediator significantly correlated with any IHC marker. In smaller numbers there was a marginal correlation (r = 0.34; P = 0.08) for epithelial tryptase mRNA with BAL fluid tryptase levels, but not with PGD2.
PGD2 and MCTC/MCTot Are Predictors of Severe Asthma
Univariate analyses.
Relationships of MC markers with hallmarks of asthma, including FEV1% predicted, frequency of asthma-related symptoms, and history of recent asthma exacerbations, were tested. Recent exacerbations of asthma were reported by 23% of mild asthma/no ICS, 19% of mild asthma/+ICS, 15% of moderate asthma, and 75% of severe asthma subjects (overall P < 0.0001). Higher numbers of submucosal MCTot were associated with better FEV1% predicted (rs = 0.34; P < 0.001) and fewer nighttime symptoms (P = 0.01), but no relationships were detected with exacerbations or other symptoms. MCTC/MCTot and BAL fluid PGD2 inversely correlated with lung function (rs = −0.38; P < 0.001 and rs = −0.27; P = 0.005, respectively). Both markers were associated with higher levels of symptoms (all P values < 0.03; Table E1), whereas higher PGD2 was associated with the history of a recent exacerbation (as defined in Methods) (6.0 pg/ml [2.0–12.0]) compared with those without (4.0 pg/ml [2.0–5.0]; P = 0.001). Other relationships between MC markers and asthma-related outcomes were not significant (data not shown).
Multivariable logistic regression models.
Only FEV1% predicted, maximum bronchodilator reversibility, MCTC/MCTot, and BAL fluid PGD2 were associated with severe asthma in the initial univariate analyses. The four highest values of MCTC/MCTot were truncated to the fifth highest value and the square root transformation was applied to decrease the influence of extreme data points on the logistic regression coefficients. In addition, the natural log transformation was applied to BAL fluid PGD2 levels before modeling for the same purpose. Model 1 (Table 3) included 62 subjects with asthma with data for all the listed predictors. In that model, both MCTC/MCTot and BAL fluid PGD2 were significant predictors of severe asthma (P = 0.02 for both predictors), when controlling for age and race. In Model 2 (n = 107), with data imputed for the missing MCTC data to compensate for the large proportion of missing submucosal MCTC/MCTot data, age was the only significant predictor of severe asthma. Both MCTC/MCTot and PGD2 were marginally important (P = 0.095 and 0.097, respectively), but still stronger predictors than FEV1% predicted (P = 0.2).
TABLE 3.
LOGISTIC REGRESSION MODELS WITH PREDICTORS OF SEVERE ASTHMA (SEVERE VS. NOT SEVERE) AMONG SUBJECTS WITH ASTHMA ONLY
| Model 1 (n = 62) |
Model 2 (n = 107) |
Model 3 (n = 98) |
||||
|---|---|---|---|---|---|---|
| Predictor | Odds Ratio | Adjusted P Value | Odds Ratio | Adjusted P Value | Odds Ratio | Adjusted P Value |
| Baseline FEV1% predicted | 1.00 (0.96, 1.04) | 0.92 | 0.98 (0.95, 1.01) | 0.20 | 0.98 (0.94, 1.02) | 0.26 |
| Maximum reversibility | 1.04 (0.99, 1.08) | 0.093 | 1.03 (1.00, 1.07) | 0.076 | 1.05 (1.00, 1.09) | 0.0372 |
| BALF PGD2* | 3.11 (1.17, 8.26) | 0.023 | 1.78 (0.90, 3.54) | 0.097 | 1.37 (0.53, 3.53) | 0.51 |
| Age, yr | 1.07 (1.01, 1.14) | 0.031 | 1.08 (1.03, 1.14) | 0.0032 | 1.10 (1.03, 1.17) | 0.0039 |
| MCTC/MCTot* | 6.16 (1.35, 28.25) | 0.019 | 3.00 (0.82, 11.00) | 0.095 | 5.08 (1.06, 24.24) | 0.0418 |
| African American (ref: white) | 2.76 (0.71, 10.68) | 0.14 | 2.52 (0.81, 7.85) | 0.11 | 3.66 (0.62, 21.68) | 0.145 |
| Clinical center | ||||||
| Overall P value | —– | 0.0263 | ||||
| CLC | 0.40 (0.03, 4.75) | |||||
| WFU | 0.19 (0.02, 2.09) | |||||
| WIS | 0.19 (0.02, 2.09) | |||||
| WSL | 0.23 (0.01, 3.87) | |||||
| PITT (reference) | 1.00 | |||||
Definition of abbreviations: BALF = bronchoalveolar lavage fluid; CLC = Cleveland Clinic, Cleveland, OH; MCTC = both tryptase- and chymase-positive mast cells; MCTot = total mast cells; PDG2 = prostaglandin D2; PITT = National Jewish Health, Denver, CO, and University of Pittsburgh, Pittsburgh, PA; WIS = University of Wisconsin, Madison, WI; WFU = Wake Forest University, Winston-Salem, NC; WSL = Washington University, St. Louis, MO.
Model 1: all listed predictors of severe vs. not severe asthma included. Model 2: multiple imputation model with data imputed for the missing MCTC. Model 3: multiple imputation model controlling for clinical center; excludes University of Virginia and Brigham and Women's Hospital due to too few cases.
Log transformation was applied to BALF PGD2 and square root transformation was applied to MCTC/MCTot for modeling purposes.
Controlling for center.
There were highly significant differences among the centers in subjects' severity of asthma, percentages of African Americans, MCTC/MCTot, BAL fluid tryptase, and PGD2 (Table E3). When controlling for the five clinical centers that had more than one subject with severe asthma in the data set (University of Virginia and Brigham and Women's Hospital were thus excluded), age, bronchodilator reversibility, and MCTC/MCTot were significant predictors of severe asthma (P values = 0.004–0.04), whereas FEV1% predicted and PGD2 were not (P = 0.26 and P = 0.51, respectively) (Table 3, Model 3). However, caution is warranted, as the prohibitive number of predictors (11) in this model limits the interpretation of results.
DISCUSSION
These results provide robust evidence for alterations in distribution, phenotypic profile, and activity of airway MCs over a range of asthma severities. In mild asthma, the submucosal MC population is abundant and maintains a “normal” phenotype profile (low MCTC). In severe asthma, the total submucosal MC population is low, but dominated by an MCTC, a phenotype that is also uniquely observed in the epithelium. Despite reduced submucosal MC counts, BAL fluid tryptase levels in severe asthma are not lower than in other groups, perhaps reflecting the reported increased capacity for tryptase activity of an MCTC-dominant MC population (30). These MC changes are further associated with higher levels of BAL fluid PGD2, a lipid mediator relatively specific to MCs, which was increased in severe asthma as compared with the mild asthma/no ICS group. These alterations in MC phenotype and likely activation correspond to greater airway obstruction, frequency of asthma symptoms, and exacerbations. In toto, these results (from the largest research bronchoscopy study yet reported) strongly suggest a role for MC differentiation, migration, and activation in the pathobiology of severe asthma.
The loss of submucosal MCs in severe asthma appears to be primarily due to the loss of MCs of the MCT phenotype. Although MCT predominate in the expanded MC population in mild asthma/no ICS, where only 20% of the MC population are MCTC, the decrease in MCT (and hence MCTot) is already present in mild asthma treated with ICS. As MCTot decrease with greater severity of asthma, the numbers and proportion of MCTC increase (P = 0.06 and P = 0.01, respectively). In the majority of subjects with severe asthma, airway MCs were predominantly MCTC. These MCTC are reported to be both “steroid-resistant” and “T cell–independent,” suggesting a mechanism for their persistence in subjects on CS therapy (23). Alterations in the airway environment in CS-treated asthma may no longer support the MCT population, but rather support or even induce MCTC differentiation. Although it is difficult to prove whether loss of MCTot is related to CS use, the airway microenvironment associated with severe asthma appears to support this MCTC phenotype even in subjects on additional leukotriene modifiers (n = 27) or anti-IgE therapy (n = 5; data not shown).
In contrast to the submucosa, epithelial MC numbers in severe asthma are not lower. Rather, epithelial MCs persist, predominantly as an MCTC phenotype. This epithelial MCTC phenotype is rarely seen in normal subjects or subjects with milder asthma, suggesting that in severe asthma, the epithelium actively contributes to MCTC migration (or differentiation), survival, and activity. Supportive of the IHC data, epithelial levels of both tryptase and CPA3 mRNA increase in asthma, and trend analysis supports a further increase with increasing severity. Previous IHC studies have suggested a redistribution of MCs to airway smooth muscle and epithelium in mild asthma (8, 15). This is the first study to report epithelial MCTC distribution in a range of asthma severities, with MCTC increase specifically in severe asthma.
The mechanisms for this epithelial migration and differentiation of MCs remain speculative. Murine models of allergic lung inflammation induce relatively limited numbers of epithelial MCs and thus preclude compartment-by-compartment studies of changes in lung MCs or phenotype (31). Therefore, studies to understand the mechanisms behind MC phenotypic changes in association with epithelial migration generally derive from murine gastrointestinal tract parasitic models (12, 32–35). After parasite exposure, MC size, distribution, and phenotype are profoundly orchestrated from initial exposure to parasite expulsion and return to basal conditions. Transforming growth factor (TGF)-β, shown to be increased in asthmatic epithelium, appears to be critical to this homing of MCs to the intestinal epithelium in mice and development of an MCTC-like phenotype crucial for resolution of Trichinella spiralis infection (32–35). Although airway epithelial-derived stem cell factor has been shown to support an MCT phenotype in vitro, whether epithelial TGF-β could promote an epithelial MCTC in human asthma remains unknown (36, 37). However, severe asthma epithelial cells, through enhanced expression of TGF-β, in association with Th2 cytokines and IL-9, could promote development of this MCTC phenotype (38). Similarly, Th2-induced epithelial eotaxins could also lead to MC recruitment and activation (39, 40).
For MCTC in severe asthma to play a role in epithelial or other cell function, evidence for phenotype-specific activation is also needed. As noted earlier, BAL fluid tryptase does not decrease as asthma severity increases. These maintained levels despite low MCTot may suggest a high level of tryptase release from MCTC cells, consistent with reports that MCTC granules contain two to three times more tryptase than MCT (30). More interestingly, however, PGD2 levels are numerically highest in severe asthma, and significantly higher than in mild asthma/+ICS. Arachidonic acid metabolism in MCTC is reported to preferentially use the cyclooxygenase pathway (21, 41). Although basal levels of this cyclooxygenase pathway product have been reported to be nearly undetectable in mild asthma, PGD2 dramatically increases after allergen/IgE stimulation (42–44). In this study, no exogenous stimulation was applied, suggesting that in severe asthma, continuous signals (IgE or otherwise) are generating PGD2.
Epithelial MC activation also likely impacts the epithelium through effects on barrier function, epithelial turnover, and even goblet cell formation (45, 46). In addition, the presumed MC expression of PGD2 can act through receptors expressed on Th2 cells (DP1 and DP2/CRTH2 [chemoattractant-homologous receptor expressed on Th2 cells]), which could further amplify Th2 processes at mucosal/epithelial surfaces (47–49). Interest in antagonizing the effects of PGD2 through these receptors is increasing, with a recent abstract reporting efficacy of a CRTH2 antagonist in mild asthma (50). A second proof-of-mechanism study of a CRTH2 antagonist also reported positive data (press release Actelion at http://www1.actelion.com). These cumulative data provide hope that antagonism of this pathway may improve treatment of severe asthma.
In the three models used, MC-associated parameters MCTC/MCTot and BAL fluid PGD2 levels were generally stronger predictors of severe asthma than FEV1% predicted, one of the most important predictors of asthma phenotype reported in a recent cluster analysis of the SARP population (51). Submucosal eosinophils were not measured in this study due to difficulties in obtaining accurate eosinophil counts by IHC in formalin fixation/paraffin embedded tissue. However, several previous studies have not shown tissue eosinophils to predict asthma severity (4, 52). Intriguingly, MCTC numbers bore no relation to atopy or BAL measures of eosinophilic inflammation, rather predominating in subjects with symptomatic severe asthma. Whether these MC alterations could tie together some of the asthma phenotypes recently described remains to be determined (51–53).
Although the direction of changes in the epithelial mRNA and BAL fluid data strongly support the IHC data, correlations between MC phenotypes measured by one approach compared with another are modest. The reasons for this are not clear, but could include differences in sensitivity of quantitative real-time PCR versus IHC, differences between stored granule proteins versus transcriptional activation, and others. Alterations in distal lung MCs could also contribute to the lack of correlations between lavage fluid and tissue/epithelial cell measures. We previously reported an increase in MCTC in the small airways of subjects with severe asthma compared with normal subjects, which could also independently contribute to lavage fluid mediator levels (5).
The large number of subjects from seven SARP centers produced the most comprehensive bronchoscopic study in humans yet reported. This approach is possible because SARP uses standardized protocols for lung function testing, clinical/immunologic evaluation, and bronchoscopy. Thus, SARP allowed comparison of data obtained by invasive procedures from a large number of subjects with a serious medical condition. However, some limitations still exist, including large differences across centers in race, severity, and three of the MC markers of interest, which likely contributed to the loss of significance for PGD2 in a model when center is included (as well as the impact of controlling for five sites). Despite a Manual of Procedures, subtle (but potentially important) differences in processing of tissue and fluids may have contributed to site differences in MC markers.
Additionally, given the nature of SARP, not every MC measurement was performed on every subject. Although automated IHC staining improved consistency, chymase staining was suboptimal in about 20% of subjects. Double staining of MC markers could give a more accurate profile of MC phenotypes. Technically, the lack of availability of primary antibodies from different species or even of different isotypes makes these approaches challenging in large-scale studies. However, a high MCTC/MCTot in severe asthma would be predicted based on the individual markers' data, as chymase-positive MCs were only detected in similar numbers to tryptase-positive cells in severe asthma. Finally, the existence of an MC phenotype expressing only chymase by IHC, which could not be identified without colocalization studies, remains highly controversial. Although the current study also cannot confirm that PGD2 is arising from MCs, the levels produced by other cells are multifold lower (54). The similar patterns of PGD2 and MCTC profiles support an MC source.
In conclusion, this comprehensive study reports a predominance of an MCTC phenotype (by both protein and mRNA) in the airway submucosa and epithelium in severe asthma, with evidence for ongoing MC activity (as measured by BAL fluid PGD2) even in the face of high inhaled and systemic CS use. Modeling suggests this shift toward the MCTC phenotype and increased airway luminal PGD2 are both more predictive of severe asthma than traditional measures. These findings have profound implications for the role that MCs play in human asthma, particularly severe asthma, as well as for identification of new therapeutic targets.
Supplementary Material
Acknowledgments
The Severe Asthma Research Program (SARP) is a multicenter asthma research group funded by the National Heart, Lung, and Blood Institute, and consisting of the following contributors (Principal Investigators are marked with an asterisk): Brigham & Women's Hospital: Elliot Israel,* Bruce D. Levy, Michael E. Wechsler, Shamsah Kazani, Gautham Marigowda; Cleveland Clinic: Serpil C. Erzurum,* Raed A. Dweik, Suzy A. A. Comhair, Emmea Cleggett-Mattox, Deepa George, Marcelle Baaklini, Daniel Laskowski; Emory University: Anne M. Fitzpatrick, Denise Whitlock, Shanae Wakefield; Imperial College School of Medicine: Kian Fan Chung,* Mark Hew, Patricia Macedo, Sally Meah, Florence Chow; University of Iowa: Eric Hoffman,* Janice Cook-Granroth; University of Pittsburgh: Sally E. Wenzel,* Fernando Holguin, Silvana Balzar, Jen Chamberlin; University of Texas-Medical Branch: William J. Calhoun,* Bill T. Ameredes; University of Virginia: Benjamin Gaston,* W. Gerald Teague,* Denise Thompson-Batt; University of Wisconsin: William W. Busse,* Nizar Jarjour, Ronald Sorkness, Sean Fain, Gina Crisafi; Wake Forest University: Eugene R. Bleecker,* Deborah Meyers, Wendy Moore, Stephen Peters, Rodolfo M. Pascual, Annette Hastie, Gregory Hawkins, Jeffrey Krings, Regina Smith; Washington University in St. Louis: Mario Castro,* Leonard Bacharier, Jaime Tarsi; Data Coordinating Center: Douglas Curran-Everett,* Ruthie Knowles, Maura Robinson, Lori Silveira; NHLBI: Patricia Noel, Robert Smith.
Supported by National Institute of Health grants HL69116, HL69130, HL69155, HL69167, HL69170, HL69174, HL69349, HL091762, KL2RR025009, M01 RR02635, M01 RR03186, M01 RR007122–14, 1 UL1RR024153, 1 UL1RR024989, 1 UL1RR024992, 1 UL1RR025008, 1 UL1RR025011, Children's Healthcare of Atlanta Center for Developmental Lung Biology.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201002-0295OC on September 2, 2010
Author Disclosure: S.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.L.F. received $10,001–$50,000 from the National Institutes of Health (NIH) in sponsored grants as a T32 & F32 NIH training grant. S.A.A.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.C.E. received more than $100,001 from Asthmatx as an investigator for an industry-sponsored grant. E.B. received $1,001–$5,000 from Amira, $10,001–$50,000 from AstraZeneca, $5,001–$10,000 from Boehringer-Ingelheim, $5,001–$10,000 from Centocor, $5,001–$10,000 from Genentech, $10,001–$50,000 from GlaxoSmithKline, $5,001–$10,000 from Merck/Schering Plough, $5,001–$10,000 from Novartis, $5,001–$10,000 from Pfizer, and $5,001–$10,000 from Wyeth in consultancy fees; industry-sponsored grants from Boehringer Ingelheim, Centocor, GlaxoSmithKline, MedImmune, Aerovance, Genentech, AstaZeneca, Ception, Pfizer, Novartis, and Amgen; and received a patent/royalties from Novartis. E.B.'s dependent/parent holds $5,001–$10,000 from GlaxoSmithKline and $5,001–$10,000 from AstraZeneca in stock ownership or options. W.W.B. received up to $1,000 from Novartis, up to $1,000 from AstraZeneca, $1,001–$5,000 from Boehringer-Ingelheim, up to $1,000 from TEVA, and $1,001–$5,000 from GlaxoSmithKline in consultancy fees; up to $1,000 from Altair, $1,001–$5,000 from GlaxoSmithKline, $5,001–$10,000 from Merck, $1,001–$5,000 from Wyeth, up to $1,000 from Pfizer, up to $1,000 from Centocor, $1,001–$5,000 from Amgen, and $1,001–$5,000 from Johnson & Johnson in advisory board fees; $1,001–$5,000 from Merck in lecture fees; and more than $100,001 from Novartis, $5,001–$10,000 from AstraZeneca, $5,001–$10,000 from Ception, $10,001–$50,000 from MedImmune, and more than $100,001 from GlaxoSmithKline in industry-sponsored grants, and more than $100,001 from the NIH-NIAID and more than $100,001 from the NIH-NHLBI in sponsored grants. M.C. received $10,001–$50,000 from Asthmatx, $1,001–$5,000 from Schering, $1,001–$5,000 from Electrocore, and $1,001–$5,000 from BMS in consultancy fees; $5,001–$10,000 from Genentech in advisory board fees; $50,001–$100,000 from AstraZeneca, $10,001–$50,000 from Boehringer Ingelheim, $10,001–$50,000 from Pfizer, $5,001–$10,000 from Genentech, and $5,001–$10,000 from Merck in lecture fees; more than $100,001 from Asthmatx, more than $100,001 from Amgen, more than $100,001 from Centocore, more than $100,001 from Ception, and more than $100,001 from GlaxoSmithKline in industry-sponsored grants; $1,001–$5,000 from Elsevier in royalties; and $10,001–$50,000 from Pfizer, more than $100,001 from Genentech, more than $100,001 from MedImmune, more than $100,001 from Merck, and more than $100,001 from Novartis for contracted research, and more than $100,001 from the NIH, more than $100,001 from the CDC, and more than $100,001 from the ALA in sponsored grants. B.G. received $2,000 from Gilead in advisory board fees, $1,000 from Apiedon for serving as an expert witness, holds a patent from Respiratory Research for breast condensate and Airborne for breast alkalinization, and is a minority shareholder in Respiratory Research. B.G.'s institution received $10,000 from Galleon in consultancy fees and $50,000 from Galleon in industry-sponsored grants., and $100,000 from the NIH as a RO1 grant. E.I. received $1,001–$5,000 from Abbott, $1,001–$5,000 from Amgen, $1,001–$5,000 from Astellas Pharma US, Inc., up to $1,000 from BI Worldwide, $10,001–$50,000 from Genentech, $5,001–$10,000 from Icagen, Inc., $10,001–$50,000 from MedImmune, $10,001–$50,000 from Merck & Co, Inc., $1,001–$5,000 from NKT Therapeutics, $10,001–$50,000 from Novartis, $1,001–$5,000 from Ono Pharmaceuticals US, $10,001–$50,000 from PDL BioPharma, $5,001–$10,000 from Pfizer, up to $1,000 from Pulmatrix, $10,001–$50,000 from Schering Plough, $1,001–$5,000 from Sepracor, and $10,001–$50,000 from Teva Specialty Pharmaceuticals in consultancy fees, $10,001–$50,000 from Genentech, $10,001–$50,000 from Merck & Co, Inc., and $10,001–$50,000 from Novartis in lecture fees; $5,001–$10,000 from Ficksman & Conley, LLP, $1,001–$5,000 from Prince, Lobel, Glovsky & Tye, LLP, $1,001–$5,000 from Campbell, Edwards & Conroy, and $1,001–$5,000 from Diedrich & Donohue for serving as an expert witness; and $50,001–$100,000 from Aerovance, $50,001–$100,000 from Amgen, $50,001–$100,000 from Asthmatx, $50,001–$100,000 from Boheringer Ingelheim, $50,001–$100,000 from Centocor, $50,001–$100,000 from Ception Therapeutics, $50,001–$100,000 from Genentech, $50,001–$100,000 from Icagen, Inc., $50,001–$100,000 from Johnson & Johnson, $50,001–$100,000 from MedImmune, $50,001–$100,000 from Novartis, and $50,001–$100,000 from Schering Plough in industry-sponsored grants, and more than $100,001 from the NIH in sponsored grants. L.B.S. received up to $1,000 from Mast Cell Pharmaceutical Co. for serving on a scientific advisory board, $50,001–$100,000 from GlaxoSmithKline, $50,001–$100,000 from Pharming, $5,001–$10,000 from Novartis, and $5,001–$10,000 from Ception in industry-sponsored grants for clinical trials; $10,001–$50,000 from Mersana in industry-sponsored grants for lab research; $50,001–$100,000 from Phadia/VCU in royalties to VCU for tryptase assay; $1,001–$5,000 from Santa Cruz/VCU, $1,001–$5,000 from Millipore/VCU, and BioLegend/VCU in royalties to VCU for mAb; up to $1,000 from Up-to-Date in royalties from sales; $1,001–$5,000 from Genentech as a clinical trial; more than $100,001 from the NIH in research grants, and more than $100,001 from the Philip Morris Research Foundations in sponsored grants from June 1, 2005 to May 31, 2008. L.B.S.'s spouse/life partner received $1,001–$5,000 from Talecris and $1,001–$5,000 from Meritage in advisory board fees; $10,001–$50,000 from Merck in speaker honoraria; and $50,001–$100,000 from GlaxoSmithKline, $50,001–$100,000 from Aventis, $50,001–$100,000 from Meritage, and $50,001–$100,000 from Novartis & Ception. D.C.–E. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.G.M. received $1,001–$5,000 from the NIH in consultancy fees for the review of grant applications and more than $100,001 from the NIH in sponsored grants as the lead statistician on several grants including AsthmaNet (Pitt site) and current SARP (Pitt site). S.E.W. received $1,001–$5,000 from Amgen, $5,001–$10,000 from GlaxoSmithKline, $1,001–$5,000 from Pearl Therapeutics, $1,001–$5,000 from Merck, and $1,001–$5,000 from Genentech in consultancy fees; $5,001–$10,000 from Amira and $10,001–$50,000 from Epigenesis in advisory board fees; $50,001–$100,000 from GlaxoSmithKline, $10,001–$50,000 from Amgen, $50,001–$100,000 from MedImmune, $50,001–$100,000 from Ception, and $10,001–$50,000 from Aerovance in industry-sponsored grants; and holds stock ownership or options in Amira and Epigenesis, and more than $100,001 from the NIH and more than $100,001 from the NIAID in sponsored grants.
References
- 1.Antonicelli L, Bucca C, Neri M, De Benedetto F, Sabbatani P, Bonifazi F, Eichler HG, Zhang Q, Yin DD. Asthma severity and medical resource utilisation. Eur Respir J 2004;23:723–729. [DOI] [PubMed] [Google Scholar]
- 2.Busse WW, Banks-Schlegel S, Wenzel SE. Pathophysiology of severe asthma. J Allergy Clin Immunol 2000;106:1033–1042. [DOI] [PubMed] [Google Scholar]
- 3.Carroll NG, Mutavdzic S, James AL. Distribution and degranulation of airway mast cells in normal and asthmatic subjects. Eur Respir J 2002;19:879–885. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med 1999;160:1001–1008. [DOI] [PubMed] [Google Scholar]
- 5.Balzar S, Chu HW, Strand M, Wenzel S. Relationship of small airway chymase-positive mast cells and lung function in severe asthma. Am J Respir Crit Care Med 2005;171:431–439. [DOI] [PubMed] [Google Scholar]
- 6.Balzar S, Strand M, Rhodes D, Wenzel SE. IgE expression pattern in lung: relation to systemic IgE and asthma phenotypes. J Allergy Clin Immunol 2007;119:855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liesker JJ, Ten Hacken NH, Rutgers SR, Zeinstra-Smith M, Postma DS, Timens W. Mast cell numbers in airway smooth muscle and PC20AMP in asthma and COPD. Respir Med 2007;101:882–887. [DOI] [PubMed] [Google Scholar]
- 8.Brightling CE, Bradding P, Symon FA, Holgate ST, Wardlaw AJ, Pavord ID. Mast-cell infiltration of airway smooth muscle in asthma. N Engl J Med 2002;346:1699–1705. [DOI] [PubMed] [Google Scholar]
- 9.Irani AA, Schechter NM, Craig SS, DeBlois G, Schwartz LB. Two types of human mast cells that have distinct neutral protease compositions. Proc Natl Acad Sci USA 1986;83:4464–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beil WJ, Pammer J. In situ detection of the mast cell proteases chymase and tryptase in human lung tissue using light and electron microscopy. Histochem Cell Biol 2001;116:483–493. [DOI] [PubMed] [Google Scholar]
- 11.Irani AM, Goldstein SM, Wintroub BU, Bradford T, Schwartz LB. Human mast cell carboxypeptidase. Selective localization to MCTC cells. J Immunol 1991;147:247–253. [PubMed] [Google Scholar]
- 12.Friend DS, Ghildyal N, Austen KF, Gurish MF, Matsumoto R, Stevens RL. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J Cell Biol 1996;135:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fokkens WJ, Godthelp T, Holm AF, Blom H, Mulder PG, Vroom TM, Rijntjes E. Dynamics of mast cells in the nasal mucosa of patients with allergic rhinitis and non-allergic controls: a biopsy study. Clin Exp Allergy 1992;22:701–710. [DOI] [PubMed] [Google Scholar]
- 14.KleinJan A, McEuen AR, Dijkstra MD, Buckley MG, Walls AF, Fokkens WJ. Basophil and eosinophil accumulation and mast cell degranulation in the nasal mucosa of patients with hay fever after local allergen provocation. J Allergy Clin Immunol 2000;106:677–686. [DOI] [PubMed] [Google Scholar]
- 15.Laitinen LA, Laitinen A, Haahtela T. Airway mucosal inflammation even in patients with newly diagnosed asthma. Am Rev Respir Dis 1993;147:697–704. [DOI] [PubMed] [Google Scholar]
- 16.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA 2007;104:15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson CK, Mori M, Bjermer L, Löfdahl CG, Erjefält JS. Alterations in lung mast cell populations in patients with COPD. Am J Respir Crit Care Med 2010;181:206–217. [DOI] [PubMed] [Google Scholar]
- 18.He S, Gaça MD, McEuen AR, Walls AF. Inhibitors of chymase as mast cell-stabilizing agents: contribution of chymase in the activation of human mast cells. J Pharmacol Exp Ther 1999;291:517–523. [PubMed] [Google Scholar]
- 19.Schulman ES, Post TJ, Henson PM, Giclas PC. Differential effects of the complement peptides, C5a and C5a des Arg on human basophil and lung mast cell histamine release. J Clin Invest 1988;81:918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oskeritzian CA, Zhao W, Min HK, Xia HZ, Pozez A, Kiev J, Schwartz LB. Surface CD88 functionally distinguishes the MCTC from the MCT type of human lung mast cell. J Allergy Clin Immunol 2005;115:1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis RA, Soter NA, Diamond PT, Austen KF, Oates JA, Roberts LJ. Prostaglandin D2 generation after activation of rat and human mast cells with anti-IgE. J Immunol 1982;129:1627–1631. [PubMed] [Google Scholar]
- 22.Moulin D, Donzé O, Talabot-Ayer D, Mézin F, Palmer G, Gabay C. Interleukin (IL)-33 induces the release of pro-inflammatory mediators by mast cells. Cytokine 2007;40:216–225. [DOI] [PubMed] [Google Scholar]
- 23.Bentley AM, Hamid Q, Robinson DS, Schotman E, Meng Q, Assoufi B, Kay AB, Durham SR. Prednisolone treatment in asthma. Reduction in the numbers of eosinophils, T cells, tryptase-only positive mast cells, and modulation of IL-4, IL-5, and interferon-gamma cytokine gene expression within the bronchial mucosa. Am J Respir Crit Care Med 1996;153:551–556. [DOI] [PubMed] [Google Scholar]
- 24.Fajt M, Wenzel SE, Balzar S. Altered levels and phenotype of intraepithelial mast cells in severe asthma [abstract]. J Allergy Clin Immunol 2009;123:S214. [Google Scholar]
- 25.Balzar S, Comhair S, Erzurum S, Wenzel SE, and SARP Investigators. Airway intraepithelial mast cells acquire a different phenotype profile in severe asthma [abstract]. Am J Respir Crit Care Med 2009;179:A3722. [Google Scholar]
- 26.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med 2000;162:2341–2351. [DOI] [PubMed] [Google Scholar]
- 27.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, et al.; National Heart, Lung, Blood Institute's Severe Asthma Research Program. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol 2007;119:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chibana K, Trudeau JB, Mustovich AT, Hu H, Zhao J, Balzar S, Chu HW, Wenzel SE. IL-13 induced increases in nitrite levels are primarily driven by increases in inducible nitric oxide synthase as compared with effects on arginases in human primary bronchial epithelial cells. Clin Exp Allergy 2008;38:936–946. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz LB, Bradford TR, Rouse C, Irani AM, Rasp G, Van der Zwan JK, Van der Linden PW. Development of a new, more sensitive immunoassay for human tryptase: use in systemic anaphylaxis. J Clin Immunol 1994;14:190–204. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz LB, Irani AM, Roller K, Castells MC, Schechter NM. Quantitation of histamine, tryptase, and chymase in dispersed human T and TC mast cells. J Immunol 1987;138:2611–2615. [PubMed] [Google Scholar]
- 31.Yu M, Tsai M, Tam SY, Jones C, Zehnder J, Galli SJ. Mast cells can promote the development of multiple features of chronic asthma in mice. J Clin Invest 2006;116:1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight PA, Wright SH, Brown JK, Huang X, Sheppard D, Miller HR. Enteric expression of the integrin alpha(v)beta(6) is essential for nematode-induced mucosal mast cell hyperplasia and expression of the granule chymase, mouse mast cell protease-1. Am J Pathol 2002;161:771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JK, Knight PA, Pemberton AD, Wright SH, Pate JA, Thornton EM, Miller HR. Expression of integrin-alphaE by mucosal mast cells in the intestinal epithelium and its absence in nematode-infected mice lacking the transforming growth factor-beta1-activating integrin alphavbeta6. Am J Pathol 2004;165:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight PA, Wright SH, Lawrence CE, Paterson YY, Miller HR. Delayed expulsion of the nematode Trichinella spiralis in mice lacking the mucosal mast cell-specific granule chymase, mouse mast cell protease-1. J Exp Med 2000;192:1849–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence CE, Paterson YY, Wright SH, Knight PA, Miller HR. Mouse mast cell protease-1 is required for the enteropathy induced by gastrointestinal helminth infection in the mouse. Gastroenterology 2004;127:155–165. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh FH, Sharma P, Gibbons A, Goggans T, Erzurum SC, Haque SJ. Human airway epithelial cell determinants of survival and functional phenotype for primary human mast cells. Proc Natl Acad Sci USA 2005;102:14380–14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu HW, Balzar S, Seedorf GJ, Westcott JY, Trudeau JB, Silkoff P, Wenzel SE. Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. Am J Pathol 2004;165:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol 2008;9:1341–1346. [DOI] [PubMed] [Google Scholar]
- 39.Bochner BS, Schleimer RP. Mast cells, basophils, and eosinophils: distinct but overlapping pathways for recruitment. Immunol Rev 2001;179:5–15. [DOI] [PubMed] [Google Scholar]
- 40.Price KS, Friend DS, Mellor EA, De Jesus N, Watts GF, Boyce JA. CC chemokine receptor 3 mobilizes to the surface of human mast cells and potentiates immunoglobulin E-dependent generation of interleukin 13. Am J Respir Cell Mol Biol 2003;28:420–427. [DOI] [PubMed] [Google Scholar]
- 41.Boyce JA. Mast cells and eicosanoid mediators: a system of reciprocal paracrine and autocrine regulation. Immunol Rev 2007;217:168–185. [DOI] [PubMed] [Google Scholar]
- 42.Murray JJ, Tonnel AB, Brash AR, Roberts LJ II, Gosset P, Workman R, Capron A, Oates JA. Release of prostaglandin D2 into human airways during acute antigen challenge. N Engl J Med 1986;315:800–804. [DOI] [PubMed] [Google Scholar]
- 43.Wenzel SE, Fowler AA III, Schwartz LB. Activation of pulmonary mast cells by bronchoalveolar allergen challenge. In vivo release of histamine and tryptase in atopic subjects with and without asthma. Am Rev Respir Dis 1988;137:1002–1008. [DOI] [PubMed] [Google Scholar]
- 44.Wenzel SE, Westcott JY, Smith HR, Larsen GL. Spectrum of prostanoid release after bronchoalveolar allergen challenge in atopic asthmatics and in control groups. An alteration in the ratio of bronchoconstrictive to bronchoprotective mediators. Am Rev Respir Dis 1989;139:450–457. [DOI] [PubMed] [Google Scholar]
- 45.Knight PA, Brown JK, Pemberton AD. Innate immune response mechanisms in the intestinal epithelium: potential roles for mast cells and goblet cells in the expulsion of adult Trichinella spiralis. Parasitology 2008;135:655–670. [DOI] [PubMed] [Google Scholar]
- 46.Guilarte M, Santos J, de Torres I, Alonso C, Vicario M, Ramos L, Martínez C, Casellas F, Saperas E, Malagelada JR. Diarrhoea-predominant IBS patients show mast cell activation and hyperplasia in the jejunum. Gut 2007;56:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagata K, Tanaka K, Ogawa K, Kemmotsu K, Imai T, Yoshie O, Abe H, Tada K, Nakamura M, Sugamura K, et al. Selective expression of a novel surface molecule by human Th2 cells in vivo. J Immunol 1999;162:1278–1286. [PubMed] [Google Scholar]
- 48.Nagata K, Hirai H, Tanaka K, Ogawa K, Aso T, Sugamura K, Nakamura M, Takano S. CRTH2, an orphan receptor of T-helper-2-cells, is expressed on basophils and eosinophils and responds to mast cell-derived factor(s). FEBS Lett 1999;459:195–199. [DOI] [PubMed] [Google Scholar]
- 49.Pettipher R. The roles of the prostaglandin D(2) receptors DP(1) and CRTH2 in promoting allergic responses. Br J Pharmacol 2008;153:S191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes NB, Pavord IP, Chuchalin AC. A randomised, double-blind, placebo-controlled study of the CRTH2 antagonist OC000459 on moderate persistent asthma. Eur Respir J 2009;34:A3144. [DOI] [PubMed] [Google Scholar]
- 51.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D‘Agostino R Jr, Castro M, Curran-Everett D, Fitzpatrick AM, et al.; National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. Identification of asthma phenotypes using cluster analysis in the severe asthma research program. Am J Respir Crit Care Med 2010;181:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol 2004;113:101–108. [DOI] [PubMed] [Google Scholar]
- 53.Haldar P, Pavord ID, Shaw DE, Berry MA, Thomas M, Brightling CE, Wardlaw AJ, Green RH. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med 2008;178:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanaoka Y, Urade Y. Hematopoietic prostaglandin D synthase. Prostaglandins Leukot Essent Fatty Acids 2003;69:163–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.