Abstract
Cat fleas, Ctenocephalides felis (Bouché) (Siphonaptera: Pulicidae), are common ectoparasites of companion animals that negatively impact their hosts directly by causing dermatitis and blood loss during feeding and indirectly through the potential transmission of disease causing agents. We isolated and characterized seven novel microsatellite loci from a partial genomic library of the cat flea enriched for di-, tri-, and tetranucleotide repeats. We screened these loci in cat fleas from two laboratory colonies and one wild-caught population collected at a temporary animal shelter (Parker coliseum) in Baton Rouge, LA. Six loci were polymorphic, with two to 15 alleles per locus and an average observed heterozygosity of 0.21 across populations. Although the two laboratory cat flea colonies were isolated from each other for many years, they did not significantly differ in their genotypic composition. The cat flea population from Parker coliseum was genetically different from the laboratory colonies, but also showed high degrees of inbreeding. Multilocus genotypes of the polymorphic loci were sufficient to assign over 85% of cat fleas to their population of origin. Genetic markers for flea population identity will allow further studies to examine the origins and movement of cat fleas with important genetic traits such as insecticide resistance or pathogen susceptibility. The use of microsatellites also could determine if there are host-specific strains of cat fleas and add insight into the development of the different subspecies of C. felis.
Keywords: genetic marker, inbreeding, cat fleas, Ctenocephalides felis
The cat flea, Ctenocephalides felis (Bouché) (Siphonaptera: Pulicidae), is a common pest of dogs and cats worldwide. The cat flea is the most important cause of flea allergic dermatitis in both dogs and cats and is also the primary intermediate host for the tapeworm Dipylidium caninum (Rust and Dryden 1997, Pugh 1987). Pet owners in the United States alone spend >US$1 billion annually on flea control products primarily used for cat flea infestations (Rust and Dryden 1997).
The cat flea also is a vector of the agents of certain diseases. Epidemiological studies on the risk factors associated with cat scratch disease established a possible role for fleas in the transmission of Bartonella hensealae, considered to be the causative agent of cat scratch fever (Zangwill et al. 1993). Subsequently, Foil et al. (1998) demonstrated experimental infection of cats with B. hensealae after inoculation of cat flea feces. Cat fleas also are competent vectors of Rickettsia felis, the etiological agent of a typhus-like illness in humans (Schriefer et al. 1994). Although it is known that R. felis can be maintained vertically in cat flea colonies (Azad et al., 1992; Wedincamp and Foil 2002), it is less clear how the bacteria move between populations of fleas. Although a vertebrate isolate has not been generated, molecular evidence suggests that R. felis is transmitted to vertebrate hosts during feeding (Wedincamp and Foil 2000). However, horizontal transmission of R. felis between fleas has yet to be demonstrated. A barrier to understanding the movement of rickettsial pathogens in flea populations is the inability to differentiate flea populations (uninfected and infected) used in horizontal transmission studies.
The goal of this study was to isolate and characterize genetic markers that would provide a comprehensive view of the population genetics of the cat flea and a method to assign fleas to their colony of origin. Microsatellites are widely recognized as the most popular genetic markers for a diversity of population and evolutionary genetic applications. Their high mutation rate and Mendelian mode of inheritance makes them especially useful to study fine-scale population genetic structure, such as genetic diversity, degree of inbreeding of populations, gene flow and migration rates, as well as assignment to populations of origin (Goldstein and Schlotterer 1999). The ability to distinguish cat flea populations will facilitate bioassays to monitor the horizontal transmission of pathogens between fleas and consequently assist in the deciphering of the ecology of cat flea-borne diseases.
Materials and Methods
Flea Collections and DNA Extraction
Three populations of C. felis were used for the studies: two colonized flea populations and one wild-caught. For colonized flea populations, newly emerged, unfed adult C. felis were either purchased from Elward II (EL) (Soquel, CA) or obtained from the Louisiana State University (LSU) colony. The EL colony is maintained on sheep blood via an artificial feeding system (Wade and Georgi 1988). The LSU colony is maintained on domestic short hair cats at the School of Veterinary Medicine in a manner described by Henderson and Foil (1993). Both colonies have a common root in that fleas maintained by J. Georgi (FleaData, Inc., Freeville, NY) were either used to supplement (EL) or to initiate (LSU) the colonies. In the past two decades, the EL colony managers have added wild-caught fleas or commercially available fleas to the colony population. In approximately the same time frame, the LSU colony has not been supplemented with outside fleas.
Hurricane Katrina passed east of New Orleans, LA, on 28 August 2005. The Parker Coliseum on the LSU campus was one of many shelters created to accommodate the pets evacuated from South Louisiana until their owners could arrange stable housing. The Parker Coliseum shelter was staffed by volunteers from the LSU community and eventually other volunteers. Protocols were gradually established by the volunteer command group and upon arrival, dogs and cats were treated for fleas as a standard procedure. From late August through October 2005, >2,000 pets were temporarily housed at the shelter within the coliseum. In the later weeks, large dogs were housed in adjacent horse stalls with dirt floors and bedding. The shelter personnel contacted L.D.F. for help with an explosive flea outbreak, presumably originating after cats from another animal shelter were admitted without being treated for fleas. In the period when arrangements were being made to treat the facility (shipment of insecticides donated by the Y-Tex Corp., Laramie, WY), L.D.F. made several flea collections from the animal stalls where dogs were previously housed by using himself as bait. There were no animals present during the collections, and all specimens were presumed to be unfed.
Specimens were stored in 95% ethyl alcohol until extraction of DNA. For the creation of a microsatellite enriched partial genomic library, 4.13 µg of high-quality genomic DNA was extracted from a pool of 60 headless fleas from a laboratory colony of the LSU School of Veterinary Sciences using the DNeasy tissue kit (QIAGEN, Valencia, CA). The heads were removed before DNA extraction to test for R. felis infection. Only DNA from Rickettsia-free fleas was included in the library. For population genetic analyses, DNA was extracted from individual fleas from the two laboratory colonies (EL and LSU) and from the population from Parker Coliseum. DNA concentrations were determined with a ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE).
Microsatellite Development
Microsatellite development essentially followed the protocol described by Booth et al. (2008), with some minor modifications. Genomic DNA fragments were ligated to SNX-linkers and enriched for loci with repeat motifs (microsatellites) by using biotinylated oligonucleotide probes with dimer, trimer, and tetramer tandem repeats (Perera et al. 2007). After capture of the of the microsatellite-enriched fragments via streptavidin-coated magnetic beads (Hamilton et al. 1999), and polymerase chain reaction (PCR) amplification with the SNX forward primer (Perera et al. 2007), enriched sequences were cloned into Escherichia coli by using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to manufacturer’s protocols.
In total, 96 clones containing inserts with a size range of 450–800 bp were sequenced at the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) by using the universal M13 forward primer. By using the Primer3Plus software (available at SourceForge.net), primer pairs were designed for 12 loci that contained at least five tandem repeats. The primer pairs were synthesized by Eurofins MWG Operon (Huntsville, AL), with the forward primer end-labeled with a M13 forward tail (Vargo and Henderson 2000).
Initially, gradient PCR reactions (with annealing temperatures ranging from 55 to 65°C) were run for each primer pair with DNA from at least three individual fleas. Of the 12 primer pairs, seven yielded scorable products. All primers worked with an annealing temperature of 57°C and uniform PCR conditions, which facilitates high-throughput genotyping. After optimization the PCR reaction mixes contained 20–40 ng of DNA template, 1× PCR buffer, 0.2 µg/µl bovine serum albumin, 2.0 mM MgCl2, 0.2 mM dNTPs, 0.4 U of Taq polymerase, 0.32 pmol of M13 F-29-IRD700 or 800, and 2.0 pmol of each primer and Millipore water (Billerica, MA) to a total volume of 10 µl.
All loci were amplified on a PTC-200 thermal cycler (MJ Research, Watertown, MA) by using a touchdown program: initial denaturation step at 94°C(30 s), followed by three cycles at 94°C (30 s), 60–1°C/cycle (30 s) and 72°C (30 s), and then 30 cycles at 94°C (30 s), 54°C (30 s), and 72°C (30 s), with a final extension step at 72°C (3 min). After adding 5 µl of Blue Stop dye solution, the amplified DNA was denatured at 95°C for 5 min and subsequently loaded onto a 6.5% 1 × Tris borate-EDTA polyacrylamide gel, and run in a LI-COR 4300 automated DNA analyzer with size standard IRDyes of 50–350 or 50–700 bp (LI-COR Biosciences, Lincoln, NE). Microsatellite allele sizes were scored using the program GeneProfiler software (Scanalytics, Inc., Fairfax, VA).
General Statistics
Numbers of alleles per locus, observed and expected heterozygosity were calculated for each locus and population using the program GDA (Lewis and Zaykin 2000). Observed and expected heterozygosities were tested for significant differences using SPSS 17.0 for Windows (SPSS GmbH, Chicago, IL). Genetic diversity was calculated using the sample-size independent rarefaction analysis of allelic richness as implemented in the FSTAT software package (Goudet 2001). To test for significance of association between genotypes of all pairs of polymorphic loci, randomized data sets (500 permutations) were created using FSTAT. Genotypes at each pair of loci were combined at random and the log-likelihood ratio G-statistic was calculated for each randomized data set. The value of P was estimated from the proportion of permutations that were greater or the same as the observed.
Population Genetic Structure
Samples from the three populations were tested for significant genotypic differentiation using log-likelihood G-statistics (FSTAT). P values were obtained through permutations of the multilocus genotypes between each pair of samples and standard Bonferroni corrections were applied. To analyze the population genetic structure and inbreeding at the levels of the individuals and populations, F-statistics were calculated using the methods of Weir and Cockerham (1984) as implemented in FSTAT or GDA, with FIS representing the standard coefficient of inbreeding in individuals relative to the population and FST representing the genetic differentiation among populations. SE values were calculated by jackknifing over loci. Finally, individuals were probabilistically assigned to genetic clusters based on their multilocus genotypes by using STRUCTURE 2.3 (Pritchard et al. 2000). Simulations used 100,000 Markov Chain Monte Carlo iterations in the burning phase and data collection phase and the default settings of the program. Separate simulations were run for models with different numbers of assumed genetic clusters (K = 2–5). Three independent runs were performed for each K to ensure consistency of the estimated posterior probabilities of K. The number of genetic clusters was determined based on the model with the highest posterior probability. Then, the estimated membership coefficients of each individual multilocus genotype was plotted to test if the genetic cluster it was assigned to was congruent with actual population of origin.
Results and Discussion
Microsatellite Characteristics
Each flea yielded, on average, 752 ng (SD = 375 ng) of DNA, which is sufficient to run ≈20–30 genotyping reactions by using the same individual. Seven microsatellite loci amplified consistently and results were repeatable. Of those loci, three had tetramer repeat units, two had trimers, and two had dimers (Table 1). Two loci consisted of compounds of different repeat sections (F2-8D, F4-3D), one was a pure but interrupted microsatellites (F4-1F), and three loci contained pure microsatellites (F4-7F, F2-10A, F3-3H) (Table 1). The repeat motif of locus F4-1F was comparatively short. Nevertheless, the locus was polymorphic and in Hardy—Weinberg equilibrium (see below).We included this locus because even short and interrupted repeat motifs have been found useful for microsatellite genotyping in other species (e.g., Ma and Yu 2009).
Table 1.
Characteristics of seven microsatellite loci developed for the cat flea
| Locus | Repeat motif | Primer sequences (5′-3′) |
Tm (°C) |
NGT | Size (bp) |
NA | He | Ho | GenBank accession |
|---|---|---|---|---|---|---|---|---|---|
| F2-8D | (ACT)3(CCT)(ACT)3(ATT)(ACT)6ACC(ACT)8 | F:gccttgttacaaacccggta | 59.9 | Lab: 60 | 188–245 | 6 | 0.44 | 0.23 | GU784825 |
| R:cacgtgacacccctctatcc | 60.4 | Parker: 33 | 4 | 0.47 | 0.21 | ||||
| F4-3D | (ACTT)2 (ACAT)2 (ACTT)(ACAT)(ACTT)13 | F:tagtgcgtttgttgcctcac | 59.9 | Lab: 56 | 172–176 | 2 | 0.12 | 0.13 | GU784827 |
| (ACTC)2(ACTT)3(ATTT)(ACTT)3 | R:aagaaagactctgctgcatgg | 59.6 | Parker: 35 | 1 | 0.00 | 0.00 | |||
| F4-5B | (CA)5 … (CA)2 … (CA)2A(CA)3ACACT(CA)2 | F:agggcctcctctcattcact | 60.2 | Lab: 55 | 141–165 | 2 | 0.15 | 0.16 | GU784828 |
| R:ggggacatggatgttttctg | 60.2 | Parker: 29 | 2 | 0.03 | 0.03 | ||||
| F4-7F | (GTTT)2 … (GTTT)6 | F:gccttgctagcagaagccta | 59.9 | Lab: 52 | 165–317 | 6 | 0.75 | 0.44 | GU784829 |
| R:gtaccgagctcgaatccact | 59.3 | Parker: 36 | 6 | 0.44 | 0.39 | ||||
| F4-1F | (TG)2 TTC(TG)3 | F:ctgcatgttgattcgtccag | 60.3 | Lab: 56 | 170–182 | 2 | 0.12 | 0.09 | GU784826 |
| R:tagcaccgcaagttatgtcg | 59.9 | Parker: 33 | 1 | 0.00 | 0.00 | ||||
| F2-10A | (GTCT)7 | F:gcatttgtatgtaacgccaaga | 60.0 | Lab: 58 | 207–283 | 15 | 0.60 | 0.35 | GU784830 |
| R:gatcgtctttgagccgaaac | 59.8 | Parker: 25 | 14 | 0.87 | 0.40 | ||||
| F3-3H | (CAG)5 | F:gcagaagcctgtggtggtag | 60.9 | Lab: 55 | 470 | 1 | 0.00 | 0.00 | GU784831 |
| R:ctcaggggtggtcactgttt | 60.0 | Parker: 32 | 1 | 0.00 | 0.00 |
Tm, melting temperature; NGT, number of individuals genotyped per locus and population (Lab, Parker); NA, number of alleles per locus and population; He, expected heterozygosity; Ho, observed heterozygosity; F:, forward; and R:, reverse. Data sets of the two laboratory populations were combined because they did not show genetic differences. Repeat motifs are in bold letters.
Population Genetic Analysis
Six of the seven microsatellite loci were polymorphic, with two to 15 alleles observed per population (Table 1). We included the monomorphic locus in the table because it consistently amplified and there is a good chance that additional alleles will be discovered when more diverse populations are genotyped in the future. None of the 15 pairs of polymorphic loci showed significant linkage (P > 0.0033, which is the adjusted P value for 5% nominal level). Thus, all six polymorphic microsatellite loci assorted independently.
Mean observed heterozygosity for all loci across all populations was 0.21 (SE = 0.06), and did not significantly differ from the expected heterozygosity 0. 37 (SE = 0.13, P = 0.12, Z = −1.572; two-tailed Wilcoxon test). We tested each locus for heterozygote excess or deficit, i.e., deviations from Hardy—Weinberg equilibrium. None of the loci showed significant heterozygote excess. Locus F4-7 showed heterozygote deficiency in the two laboratory populations, and loci F2-8D and F2-10A showed heterozygote deficit in both the laboratory populations and the population collected from Parker Coliseum (P = 0.0028; adjusted nominal level for 5% significance after Bonferroni correction).
Permutation tests of the distribution of genotypes between pairs of flea populations showed no significant differentiation between the two laboratory colonies and Nei’s unbiased identity index was >0.99 (Nei 1978). Therefore, the data sets of the laboratory populations were combined. The lack of genetic differentiation between the laboratory colonies confirmed their common origin. Apparently, the cat fleas that were introduced into the EL colony since the LSU colony was established did not change the genetic profile, either because the introduced and colony fleas were genetically similar or the introduced fleas did not sufficiently reproduce in the colony.
However, the population from animal stalls at Parker coliseum differed significantly from the combined laboratory populations in the permutation test, and both Nei’s identity (0.91) and FST (0.15) indicate moderate genetic distance. The combined laboratory population had five private alleles with >5% frequency in the population that distinguished it from the Parker Coliseum population. The latter population had two private alleles with >5% frequency across all six polymorphic loci. F-statistics showed high levels of inbreeding in both, the laboratory and the Parker Coliseum populations (Lab: FIS = 0.24 [SE = 0.13], Parker: FIS = 0.30 [SE = 0.15]), and moderate allelic richness (Lab: 4.42 [SD = 1.38], Parker: 4.51 [SD = 2.02]) across all polymorphic loci. Although the laboratory populations were largely isolated from each other for two decades, which explains the heterozygote deficiency and lack of genetic diversity due to repeated inbreeding cycles, the high levels of inbreeding in the Parker coliseum population was surprising.
Although the laboratory population and the wild-caught population showed only moderate genetic diversity, the presence of private alleles and differences in allele frequencies between the populations was sufficient to assign the majority of individuals to their population of origin with high confidence (fig. 1). When all polymorphic loci were included initially, Bayesian simulations using the program STRUCTURE indicated the existence of three distinct genetic clusters. Individuals within the combined laboratory populations seemed to be assigned in about equal proportions to two genetic clusters, which were not congruent with the origin from the EL or LSU laboratory populations. Recent introduction of new fleas to both of the laboratory populations would be unlikely due to the isolation of the laboratory colonies from each other. At this point we have no evidence that this substructure is of biological significance (e.g., indicating assortative mating, sex linked loci). Furthermore, this substructure is based on only one locus (F2-10A) and not supported by the other five loci. Despite the perceived substructure in the combined laboratory population, we still were able to correctly assign 49 of 52 multilocus genotypes from the combined laboratory colonies and 22 of the 25 genotypes from the Parker population (with memberships coefficients >50%).
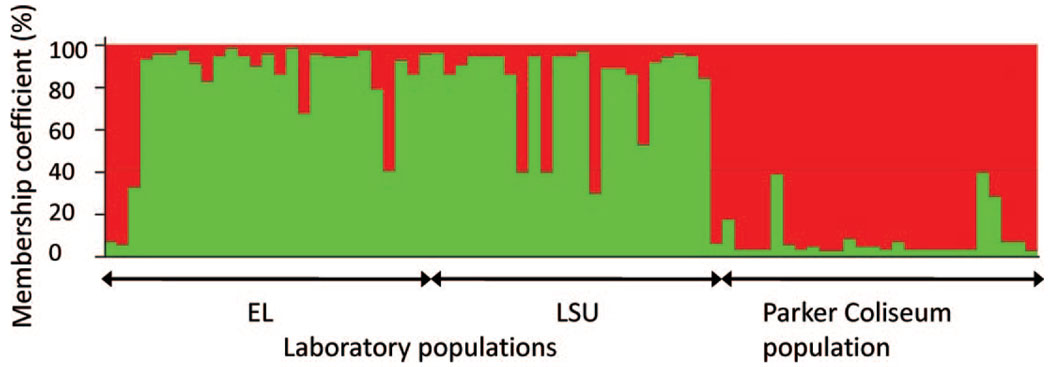
Fig. 1.
Assignment of individual multilocus genotypes to three genetic clusters inferred from STRUCTURE simulations. Genotypes originated from two laboratory populations (1, EL; 2, LSU) and one population collected from an animal stall (3, Parker Coliseum). Each bar represents the membership coefficient, Q, divided into parts proportional to proposed ancestry of an individual genotype in each of the three genetic clusters. (Online figure in color.)
When locus F2-10A was omitted from the data set, Bayesian simulations indicated only two genetic clusters, and the genetic cluster individual genotypes were assigned to was largely congruent with their populations of origin, i.e., the combined laboratory population versus the population from Parker coliseum (fig. 1). The majority (>85%) of genotypes were assigned strongly (with a membership coefficient >80%) to their own population. Seven genotypes from the laboratory population and three from the Parker population showed an ambiguous assignment (membership coefficient between 40 and 60%). Only three genotypes of the laboratory population and none of the Parker population were assigned wrong (with a membership coefficient of >80%). Thus, the origin of the large majority of individuals can be assigned to their population of origin based on multilocus genotypes even if the populations are only moderately differentiated and show a rather low genetic diversity (high levels of inbreeding).
Initially, we expected that the Parker collection would represent diverse populations of cat fleas from many locations and animals. The lack of genetic diversity of the fleas collected at the shelter is probably associated with a single introduction of fleas. Although we cannot absolutely confirm the notion that the flea infestation was caused by a single introduction, the microsatellite data would not contradict that assumption. Furthermore, all of the fleas collected at the shelter were in the horse stalls and were C. felis. The open coliseum where the majority of the pets were housed in cages was not infested. An infestation of 100 fleas on cats can go undetected without careful examination, meaning that >1,000 eggs per day are often produced when cats are not protected. Female fleas can survive for at least 50 d if they escape grooming, and when feeding on cats can produce >25 eggs per day, with 50% adult emergence occurring 20 d later (Moser et al. 1991, Rust and Dryden 1997). After 20 d, a single female’s progeny of half brothers and sisters from 1 d could produce 300 eggs per day or a week’s worth of progeny could produce 2,100 eggs. After 40 d with available hosts, an environment could have thousands of cat fleas descended from the same mother.
If there are populations of cat fleas with signature microsatellites, then genetic profiling could be used to trace origins of infestations. Flea eggs are laid and fall off the host and the larvae develop on the feces of the feeding adults. Introductions into households, e.g., are made when the companion animal ranges into an area where fleas are present and returns home. The use of microsatellites to compare infestations will probably be revealing regarding flea population movement. If future studies support that application, then tracing the origins and movement of fleas with important genetic traits such as insecticide resistance or pathogen susceptibility would be possible.
The use of microsatellites could help determine whether there are host-specific strains of cat fleas. Cat fleas are known to infest many different mammalian hosts including cats, dogs, horses, sheep, poultry, and several wild hosts (Rust and Dryden 1997). Williams (1993) found that cat fleas reared from fleas collected from areas with infested cats had a reduced egg viability of 23–33% when they were fed on calves rather than cats. Dryden et al. (1993) reported a cat flea infestation of dairy calves that had lasted for at least 5 yr or up to 60 generations, and fleas collected from the dairy could not be maintained on cats. Microsatellite markers could be used to test whether there are genetically different cat fleas separated by host species.
Furthermore, comparing the genotypic profiles of cat fleas collected from different continents could provide insight into the development of different subspecies: According to the Catalog of Life 2007 annual checklist (Catalog of Life 2007), three subspecies of the cat flea are recognized: Ctenocephalides felis felis (Bouché, 1835), Ctenocephalides felis damarensis Jordan, 1936, and Ctenocephalides felis strongylus (Jordan 1925). Lewis (1972) indicated that C. f. felis was cosmopolitan in distribution, whereas C. f. damarensis and C. f. strongylus were found in the East African Subregion. Lewis (1972) also listed Ctenocephalides felis orientalis (Jordan 1925) as a subspecies found from India to Australia.
The microsatellites developed in this study will allow for detailed epidemiological studies on cat flea-borne disease-causing agents, such as R. felis. The current distribution of R. felis includes 30 countries; with a majority of the descriptions of infection in the cat flea host (reviewed in Reif and Macaluso, 2009). The incidence of R. felis infection in wild-caught fleas is highly variable, and there many colonized cat flea populations with varying (0–100%) prevalence of R. felis infection (Higgins et al. 1994). The method of introduction of R. felis into a population of fleas is unknown. Likewise, the observation that in any given survey of wild-caught or colonized cat fleas the prevalence of these vertically maintained bacteria is not ubiquitous suggests that there are factors preventing spread of R. felis through a population. Several possibilities have been put forth, including microbial competitive interaction (Reif and Macaluso 2009). Also, it is possible that genotypic variation of cat fleas results in variable susceptibility to R. felis. Up to this point there has been no tool to assess cat flea strain or population differences and, thus, no way to determine if differences in vector competence exist. Subsequent studies can use microsatellite loci identified in the current study to differentiate cat flea strains relative to the distribution of R. felis in flea populations. Ultimately, these analyses will enhance our understanding of the ecology of this emerging flea-borne pathogen.
Acknowledgments
We thank Mark Guillotte for technical assistance and Warren Booth (North Carolina State University) for advice with microsatellite development. This work was supported by the National Institutes of Health National Institutes of Allergy and Infectious Disease (AI069248). This article was approved for publication by the Director, Louisiana Agricultural Experiment Station, as manuscript 2010-234-4456
References Cited
- Azad AF, Sacci JB, Jr, Nelson WM, Dasch GA, Schmidtmann ET, Carl M. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. Proc. Natl. Acad. Sci. U.S.A. 1992;89:43–46. doi: 10.1073/pnas.89.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth W, Lewis VR, Taylor RL, Schal C, Vargo EL. Identification and characterization of 15 polymorphic microsatellite loci in the western drywood termite, Incisitermes minor (Hagen) Mol. Ecol. Res. 2008;8:1102–1104. doi: 10.1111/j.1755-0998.2008.02169.x. [DOI] [PubMed] [Google Scholar]
- Catalogue of Life. Parhost World Database of Fleas. GBIF data portal; 2007. Annual checklist. http://data.gbif.org/datasets/resource/1564. [Google Scholar]
- Dryden MW, Broce AB, Moore WE. Severe flea infestation in dairy calves. JAVMA. 1993;203:1448–1452. [PubMed] [Google Scholar]
- Foil LD, Andress E, Freeland RL, Ray AF, Rutledge R, Triche P, O’Reilly K. Experimental infection of cats with Bartonella hensalae by inoculation of cat flea (Ctenocephalides felis (Bouche) (Siphonaptera: Pulicidae) feces. J. Med. Entomol. 1998;35:625–628. doi: 10.1093/jmedent/35.5.625. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Schlotterer C. Microsatellites: evolution and applications. Oxford, United Kingdom: Oxford University press; 1999. [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3. http://www.unil.ch/izea/softwares/fstat.html.
- Hamilton MB, Pincus EL, Di Fiore A, Fleischer RC. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques. 1999;27:500–507. doi: 10.2144/99273st03. [DOI] [PubMed] [Google Scholar]
- Henderson G, Foil LD. Efficacy of Diflubenzuron in simulated household and yard conditions against the cat flea Ctenocephalides felis (Bouché) (Siphonaptera: Pulicidae) J. Med. Entomol. 1993;30:619–621. doi: 10.1093/jmedent/30.3.619. [DOI] [PubMed] [Google Scholar]
- Higgins JA, Sacci JB, Jr, Schriefer ME, Endris RG, Azad AF. Molecular identification of rickettsia like microorganisms associated with colonized cat fleas (Ctenocephalides felis) Insect Mol. Biol. 1994;3:27–33. doi: 10.1111/j.1365-2583.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D. Genetic Data Analysis: computer program for the analysis of allelic data, version 1.0 (d12) Free program distributed by the authors. http://alleyn.eeb.uconn.edu/gda/
- Lewis RE. Notes on the geographical distribution and host preferences in the order Siphonaptera. Part I. Pulicidae. J. Med. Entomol. 1972;9:511–520. doi: 10.1093/jmedent/9.6.511. [DOI] [PubMed] [Google Scholar]
- Ma H, Yu Z. Development of twenty-two polymorphic microsatellite loci in the noble scallop, Chlamys nobilis. Conserv. Genet. 2009;10:1587–1590. [Google Scholar]
- Moser BM, Koehler PG, Patterson RS. Effect of larval diet on cat flea development times and adult emergence. J. Econ. Entomol. 1991;84:1257–1261. doi: 10.1093/jee/84.4.1257. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera O, Snodgrass GL, Scheffler BE, Gore J, Abel CA. Characterization of eight polymor hic microsatellite markers in the tarnished Plant bug, Lygus lineolaris (Palisot de Beauvois) Mol. Ecol. Notes. 2007;7:987–989. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh RE. Effects on the development of Dyplidium caninum and on the host reaction to this parasite in the adult flea (Ctenocephalides felis felis) Parasitol. Res. 1987;73:171–177. doi: 10.1007/BF00536475. [DOI] [PubMed] [Google Scholar]
- Reif KE, Macaluso KR. Ecology of Rickettsia felis: a review. J. Med. Entomol. 2009;46:723–736. doi: 10.1603/033.046.0402. [DOI] [PubMed] [Google Scholar]
- Rust MK, Dryden MK. The biology, ecology, and management of the cat flea. Annu. Rev. Entomol. 1997;42:451–473. doi: 10.1146/annurev.ento.42.1.451. [DOI] [PubMed] [Google Scholar]
- Schriefer ME, Sacci JB, Jr, Dumler JS, Bullen MG, Azad AF. Identification of a novel rickettsial infection in a patient diagnosed with murine typhus. J. Clin. Microbiol. 1994;32:949–954. doi: 10.1128/jcm.32.4.949-954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo EL, Henderson G. Identification of polymorphic microsatellite loci in the Formosan subterranean termite Coptotermes formosanus Shiraki. Mol. Ecol. 2000;9:1935–1938. doi: 10.1046/j.1365-294x.2000.0090111935.x. [DOI] [PubMed] [Google Scholar]
- Wade SE, Georgi JR. Survival and reproduction of artificially fed cat fleas, Ctenocephalides felis Bouché (Siphonaptera: Pulicidae) J. Med. Entomol. 1988;25:186–190. doi: 10.1093/jmedent/25.3.186. [DOI] [PubMed] [Google Scholar]
- Wedincamp J, Jr, Foil LD. Infection and seroconversion of cats exposed to cat fleas (Ctenocephalides felis Bouché) Infected with Rickettsia felis. J. Vector Ecol. 2000;25:123–126. [PubMed] [Google Scholar]
- Wedincamp J, Jr, Foil LD. Vertical Transmission of Rickettsia felis in the Cat Flea (Ctenocephalides felis Bouché) J. Vector Ecol. 2002;27:96–203. [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams B. Reproductive success of cat fleas, Ctenocephalides felis, on calves as unusual hosts. Med. Vet. Entomol. 1993;7:94–98. doi: 10.1111/j.1365-2915.1993.tb00658.x. [DOI] [PubMed] [Google Scholar]
- Zangwill KM, Hamilton DH, Perkings BA, Regnery RL, Plikytis BD, Hadler JK, Carter ML, Wenger JD. Cat scratch disease in Connecticut. N. Engl. J. Med. 1993;329:8–13. doi: 10.1056/NEJM199307013290102. [DOI] [PubMed] [Google Scholar]