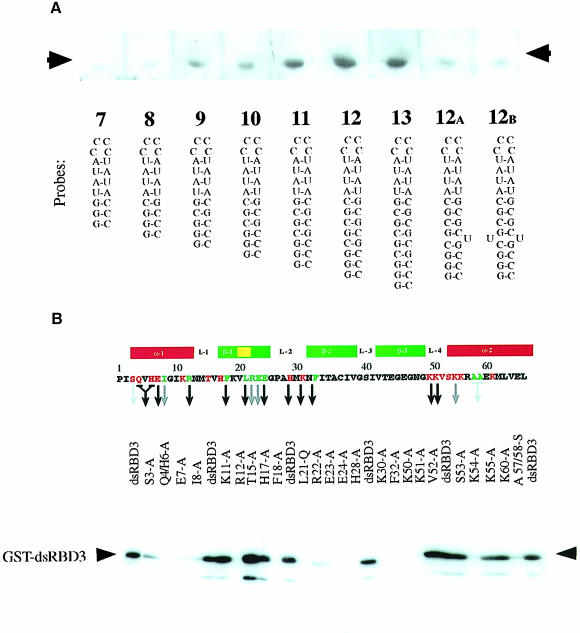
Fig. 1. Biochemical characterization of Staufen dsRBD3 interaction with RNA stem–loops. North-western blots showing (A) binding of wild-type dsRBD3 to RNA stem–loops and (B) alanine-scanning mutagenesis of dsRBD3. The top panel shows the positions of the amino acids that were substituted, with a diagram of the secondary structure of the domain underneath. The numbering refers to the general dsRBD alignment scheme; Pro1 corresponds to Pro579 in Drosophila Staufen. Green residues correspond to mutated amino acids required for the correct folding of the domain, while red residues are surface exposed. A black arrow indicates no RNA binding; grey arrow, reduced binding; no arrow indicates that the mutation had no effect. The lower panel shows a representative blot of the alanine substitution mutants probed with double-stranded VA1 RNA. The same amount of protein was present in each lane (data not shown).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.