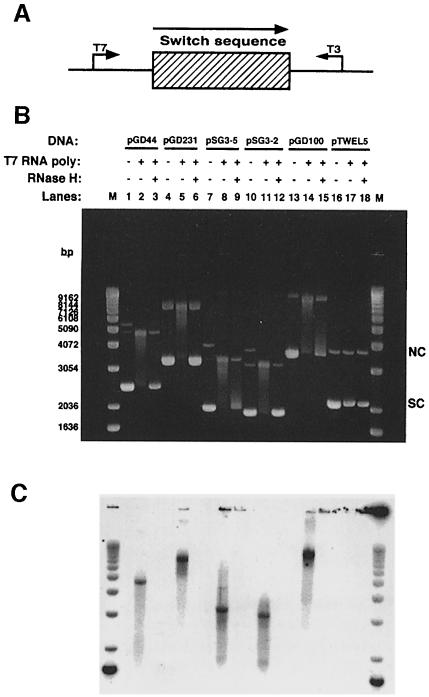
Fig. 1. Altered DNA mobility upon in vitro transcription of murine class switch sequences Sμ, Sγ3 and Sγ2b. (A) Diagram of switch sequences on plasmids showing the direction of physiological transcription. The bent arrows indicate the direction of transcription by either T3 or T7 RNA polymerase. (B) Supercoiled plasmid DNA containing either a 900 bp fragment of Sμ (pGD44), a 2.2 kb fragment of Sγ3 (pGD231), a 267 bp fragment of Sγ3 (pSG3-5), a 129 bp fragment of Sγ3 (pSG3-2), an 832 bp fragment of Sγ2b (pGD100) or a 564 bp HindIII fragment from λ phage (pTWEL5) was transcribed, treated with RNase A, run out on a 1% agarose gel and post-stained with ethidium bromide as described in Materials and methods. Lanes 1, 4, 7, 10, 13 and 16 are non-transcribed plasmids; lanes 2, 5, 8, 11, 14 and 17 are plasmids transcribed with T7 RNA polymerase; lanes 3, 6, 9, 12, 15 and 18 are plasmids transcribed with T7 RNA polymerase and treated with RNase H. A 1 kb ladder (Gibco-BRL) was used as a molecular weight marker (M). The positions of supercoiled (SC) and nicked circular (NC) forms of the plasmids are indicated. (C) Radioactive image of the gel shown in (B).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.