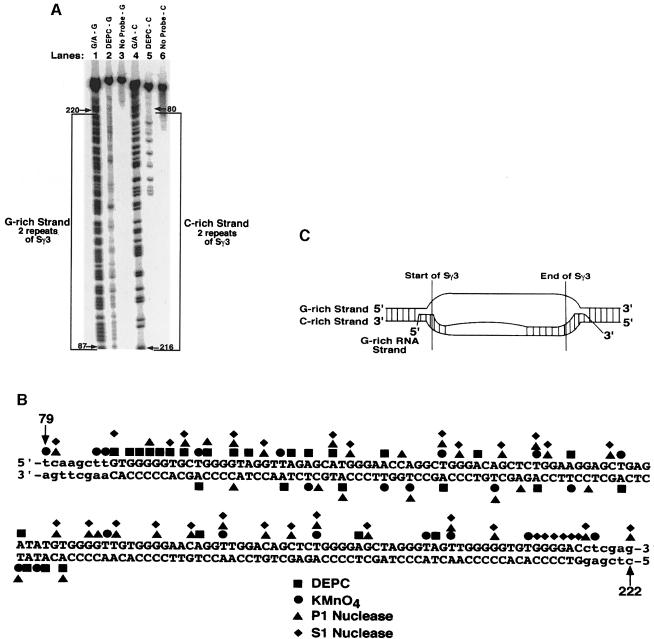
Fig. 2. Fine mapping of the chemical and enzymatic hypersensitive sites on the G–rich and C–rich DNA strands within the R–loop formed at two repeats of Sγ3. (A) Sequencing gel showing the results of probing the G–rich and C–rich DNA strands within the R–loop formed at two repeats of Sγ3 on supercoiled pSG3-2 with DEPC (lanes 2 and 5, respectively), or the absence of probing (lanes 3 and 6, respectively). The positions of modification were determined by using a G + A sequencing ladder for each strand (lane 1 for the G–rich strand and lane 4 for the C–rich strand). The G–rich and C–rich strands are bracketed. (B) Summary of fine mapping with DEPC, KMnO4, P1 nuclease and S1 nuclease. Bases in upper case letters depict the G–rich (top) and the C–rich (bottom) strands of two repeats of Sγ3, whereas the lower case letters represent the vector sequence. (C) Structure of the RNA–DNA hybrid at the two repeats of Sγ3 as inferred from the fine mapping data. The boundaries of two repeats of Sγ3, and the configuration of the DNA and RNA strands are shown.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.