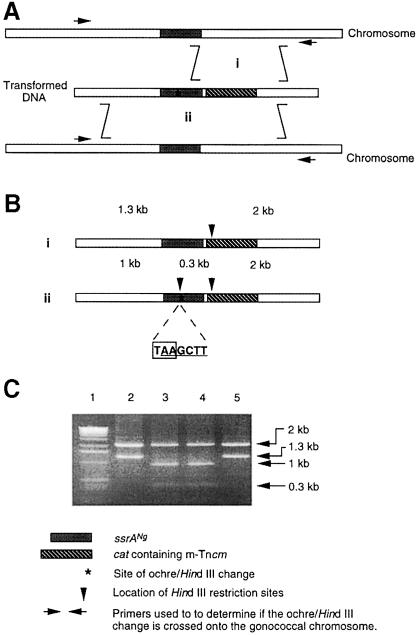
Fig. 5. Details of the allelic replacement that placed ssrANg mutant alleles on the gonococcal chromosome. Construction of a recombinant chromosomal ssrANg gene with the ochre mutation is used as an example of the procedure. (A) The double crossover recombination events resulting in allelic replacement. Selection is for Cmr carried by the linked m-Tncm. The m-Tncm will be crossed onto the chromosome without the ochre mutation if the recombination occurs as shown in (i). The ochre mutation (indicated by the asterisk) will be crossed with the m-Tncm onto the chromosome if the recombination occurs as shown in (ii). Arrows indicate the positions of the primers used in (B) and (C). (B) Schematic representation of the PCR products of the allelic replacements diagrammed in (A). The downward pointing arrows indicate the locations of the HindIII sites used in the analysis of the PCR products shown in (C). The location of the ochre codon is shown in (ii). (C) Cleavage of PCR products confirming proper allelic replacement. Cmr transformant colonies were used as the source of template DNA for the PCRs. PCR products, generated using the primers indicated in (A), were cleaved with HindIII and the products analyzed by electrophoresis in an agarose gel [source of DNA: lane 1, DNA 1 kb ladder (Gibco); lane 2, wild-type allele; lane 3, plasmid with cloned ssrANg/ochre allele; lanes 4 and 5, transformants]. Because an engineered HindIII site was positioned adjacent to the ochre mutation, PCR products from transformants that have an extra HindIII site, as shown in lane 4, are likely to carry the ochre mutation. Therefore, these transformants were chosen for sequence analysis to determine definitively if the transformant had the desired mutation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.