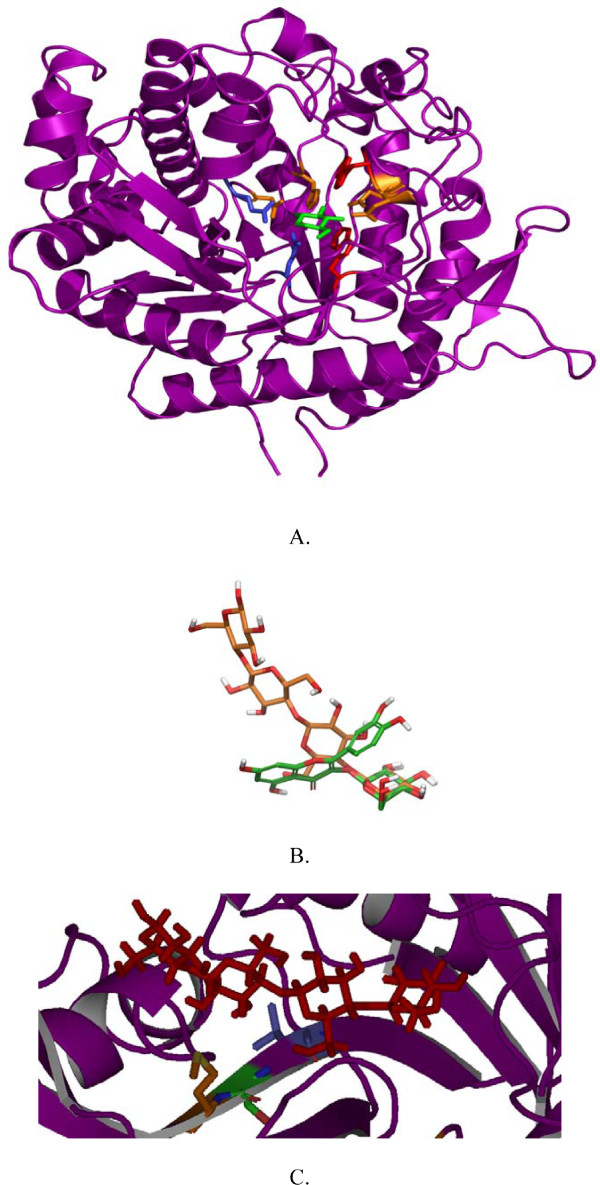
Figure 2.
Overall structure of TnBgl1A, relative substrate positions and the cellotetraose position in relation to mutated residues. TnBgl1A homology model showed the typical (β/α)8 barrel fold (A.), a feature of the overall structure in GH1. The proton donor E164 and nucleophile E349 of the enzyme are illustrated in blue and shown in stick representation. In green the G2F inhibitor is shown in the -1 subsite, interacting with four residues in orange: (Q18, H119, N163, E403) by hydrogen bonds and two by hydrophobic interactions (W396, W404). In panel B the relative positions of the ligands cellotetraose (orange) and quercetin-3-glucoside (Q3, in green) when bound in the enzyme are shown. The matching binding of the glucopyranoside in the two substrates at the -1 subsite is shown to the right. The cellotetraose labelling from left to right correspond to subsites +3, +2, +1, -1. In panel C the cellotetraose (again positioned with the +3 subsite to the left), is displayed in the active site channel, and the selected residues close to the +2 subsite (from left to right:G222, N221 and F219) are shown in the mutated forms as M222, S221 and L219. The G222M was made to increase hydrophobicity at the entrance of the active site, while the G222Q mutation (not shown) was predicted to result in hydrogen bonding with the substrate. The F219L and N221S mutations were predicted to generate space for better substrate accommodation.