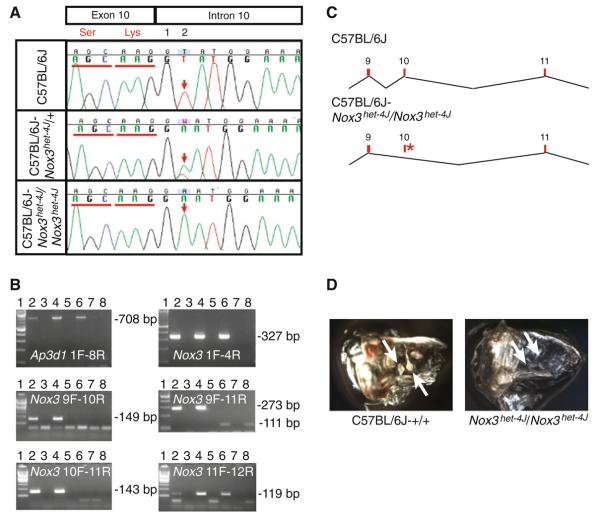
Fig. 5.
Analysis of the Nox3het–4J allele. a Sanger sequencing-based identification of the Nox3het–4J mutation. The mutation consists of a T > A transversion at the +2 position of the splice donor site just after exon 10. b RT-PCR analysis of C57BL/6J and Nox3het–4J/Nox3het–4J inner-ear RNA with Ap3d1 1F/8R (positive control) primers as well as the Nox3 1F/4R, 9F/10R, 9F/11R, 10F/11R, and 11F/12R primer pairs. Lane 1, 100-bp ladder; lane 2, wild-type RNA; lane 3, negative reverse transcriptase (−RT) control; lane 4, Nox3het–4J/+ RNA; lane 5, −RT control; lane 6, Nox3het–4J/Nox3het–4J RNA; lane 7, −RT control; lane 8, no template control. c Models of wild-type (C57BL/6J) and mutant (C57BL/6J-Nox3het–4J/Nox3het–4J) Nox3 transcripts. Red numbered rectangles, exons; asterisk, mutation. d Whole-mount preparation of cleared temporal bones from adult C57BL/6J-+/+and Nox3het–4J/Nox3het–4J mice. Note the presence of otoconia (arrows) in the wild-type preparation and their absence in mutant homozygotes