Abstract
This paper presents the mantisGRID project, an interinstitutional initiative from Colombian medical and academic centers aiming to provide medical grid services for Colombia and Latin America. The mantisGRID is a GRID platform, based on open source grid infrastructure that provides the necessary services to access and exchange medical images and associated information following digital imaging and communications in medicine (DICOM) and health level 7 standards. The paper focuses first on the data abstraction architecture, which is achieved via Open Grid Services Architecture Data Access and Integration (OGSA-DAI) services and supported by the Globus Toolkit. The grid currently uses a 30-Mb bandwidth of the Colombian High Technology Academic Network, RENATA, connected to Internet 2. It also includes a discussion on the relational database created to handle the DICOM objects that were represented using Extensible Markup Language Schema documents, as well as other features implemented such as data security, user authentication, and patient confidentiality. Grid performance was tested using the three current operative nodes and the results demonstrated comparable query times between the mantisGRID (OGSA-DAI) and Distributed mySQL databases, especially for a large number of records.
Key words: Healthgrid, digital imaging and communications in medicine (DICOM), grid computing, health level 7 (HL7), high-performance computing
Introduction
Grids have been introduced and proposed to address several challenges associated with storage, processing, and availability of data in a variety of scientific fields.1–3 Currently, huge amounts of data are generated everyday by the biomedical community including, but not limited to, medical imaging, genomics, drug discovery, treatment planning, and biomedical instruments. However, data should not only be stored and rather needs to be readily accessible and integrated either for clinical or research purposes. Furthermore, collaboration across clinical and research centers is becoming critical to address the challenges of modern medicine where teamwork and association is mandatory.
Grid technologies have emerged as a viable tool to manage large volumes of information and computational resources necessary in several areas. Also, they have proven to be a reliable and efficient distributed infrastructure for the management of medical images. A number of grid projects and developments in the biomedical area include: the European mammogram database MammoGrid,4 the biomedical platform MedIGrid,5 VirtualPACS (a grid client application for the digital imaging and communications in medicine [DICOM] DataService),6 caGrid (an open source grid software infrastructure aimed at enabling multi-institutional data sharing and analysis originated at the NCI Cancer Biomedical Informatics Grid [caBIG™]),7 and the European grid-based pediatrics platform Health-e-Child,8 among others. In addition, several research projects, focused on the augmented computing power of grids, have been reported in applications such as radiation therapy dosimetry planning through Monte Carlo simulations,1 multiple sclerosis brain segmentation,9 and tomographic image reconstruction.10
Medical imaging alone continues to create significant storage and computing power demands. Consider as an example the new emerging clinical technology of dual-energy computed tomography (CT).11 A state of the art CT scanner can be operated at 0.6 mm collimation to produce thin isotropic resolution slices; but in the dual-energy CT mode, three datasets are stored: the high (140 kV) and the low (80 kV) energy scans, plus a mixed dataset (equivalent to 120 kV conventional single-energy CT). This without taking into account the possibility of the patient undergoing both contrast and noncontrast scans, a second examination, or further postprocessing results, leading to various gigabytes of information from a single patient, and easily at least five times more information than conventional single-source, single-energy CT scans. This example and similar ones in other imaging modalities (e.g., 3D magnetic resonance imaging [MRI] T1 and T2 scans, PET/CT, etc.) provide motivation on why computational grids providing shared storage resources and increased computational power appear to be highly desirable.
Grid technologies present significant opportunities to developing countries and in particular for healthcare and telemedicine.12–14 Integration of developing countries into the global context of healthgrids is of primary importance because of the special needs of different communities and sometimes different epidemiologies due to phenotypic differences in people from different race, ethnics, diet, or even life style; in addition, such integration help developing countries’ communities to keep track and benefit from technological advances.
In Latin America in particular, there exist different initiatives in cross-collaboration with Europe to develop grid infrastructures with a variety of purposes including biomedical applications, engineering, e-learning, physics, environmental sciences, and others.14 However, among this few projects, none seem to have been strongly established for medical imaging management. On the other hand, the current availability of increasingly reliable software tools to support grids and in particular healthgrids, in many cases with free access, provides an excellent opportunity for developing countries to join the benefits of the use of grids. Such scenario of needs in Latin American communities for healthgrids and the availability of tools motivated the creation of the mantisGRID15,16 by various Colombian clinical and academic institutions.
The present article introduces the mantisGRID platform for managing and sharing of information over a grid infrastructure in the RENATA network (Colombian High Technology Academic Network). mantisGRID considers a data abstraction architecture via Open Grid Services Architecture Data Access and Integration (OGSA-DAI)17,18 middleware services and supported by the Globus Toolkit.19,20 The OGSA describes an architecture for a service-oriented grid computing environment for business and scientific use, developed within the Global Grid Forum. OGSA-DAI™ is a middleware bundle that allows data resources, such as file systems, relational, or Extensible Markup Language (XML) databases, to be accessed, federated, and integrated across the network, following the OGSA standard. These services provide integration of the information using heterogeneous and distributed database engines. Each node of the grid is autonomous in the management of users and DICOM datasets, and all nodes are interconnected via RENATA. What is special about mantisGRID is its simplicity. It was designed from the beginning as an alternative to data distribution optimizing the grid usage. Interaction with the user is accomplished via dynamic Web page using the hypertext preprocessor (PHP) language. Even though PHP is popular in Web applications, the interaction among the GRID middleware and PHP was almost nonexistent until now.
This paper summarizes the technical and clinical features developed in mantisGRID. The “mantisGRID Architecture” section discusses the architecture of mantisGRID. The “DICOM and SR-CDA HL7 Interoperability in mantisGrid” section describes the interrelation between the relational database created to manage the DICOM objects created with XML schema documents, while the “Security” section includes a description of additional features such as data confidentiality and user authentication. The “Performance TEST” section illustrates the performance of the grid over the current functional nodes located in three different institutions. The “Discussion and Future Work” section provides some discussion, conclusions, and future directions of the mantisGRID project.
MantisGRID Architecture
The mantisGRID focuses on building a pilot platform based on open source grid infrastructure. It provides services to access and exchange medical images and associated information that comply with DICOM and health level 7 (HL7) standards. mantisGRID was designed to deal with diverse medical imaging modalities such as CT, X-ray radiography, magnetic resonance, ultrasound, nuclear medicine, and visible light digital medical photography. However, the initial clinical testing has been performed for two specialties, dentistry and dermatology, by the use of panoramic X-ray radiography and visible light medical photography.
The mantisGRID uses a series of protocols and services (middleware), as well as a virtual repository of medical images that support the concept of data grid. The virtual repository database is physically located and operated by different organizations. The institutions involved in mantisGRID include both medical and academic centers which together create a virtual organization unifying their individual databases as a single resource.
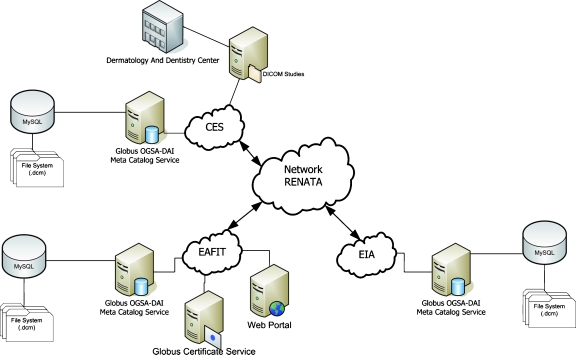
Figure 1 shows the general architecture of mantisGRID. Each node, representing an institution, consists of a Web-feeding portal and a database. The EAFIT and EIA nodes represent two academic institutions (Colombian universities), while the CES node represents both an academic institution (CES University) and also a local hospital specializing in dermatology and dentistry. Each node has a middleware tool that enables the grid technology functionality inside the architecture.
Fig 1.
General architecture of mantisGRID. The three main nodes are interconnected via the RENATA network.
The application that coordinates the image data transport from a node to the mantisGRID user is called File Trower and is present in each node. These nodes are integrated into mantisGRID through OGSA-DAI17,18 and the Globus Toolkit.19,20 The Globus Toolkit modules provide diverse services related to security and data management based on standard specifications of OGSA. OGSA-DAI is used in the integration of data on the grid. One of the nodes of the mantisGRID is denominated as the Master Node and is responsible for managing the data sources and ensures security in the databases. The software tools used in OGSA-DAI implementation are:
Globus Toolkit 4.0.5 for the deployment of Web Services Globus in OGSA-DAI.
WebApplication Tomcat for Web Services.
OGSA-DAI 3.0 for OGSA middleware implementation in Apache Tomcat containers.
OGSA Distributed Query Processing (OGSA-DQP) version 3.2.1. An OGSA-DAI component that supports queries over OGSA-DAI data resources and over other services on the grid.
With these services, mantisGRID fulfills the initial requirements related to:
Authentication/authorization: at each institution (node) level validating against its database.
DICOM Study Access: all DICOM objects are accessed at local level by the File Trower application from the file system.
DICOM Study Search, Localization, and Visualization.
Dentistry and Dermatology DICOM studies creation and DICOM Structured Reporting–Clinical Document Architecture (SR-CDA) HL7 generation.
Fulfillment of legal requirements regarding security of clinical information.
OGSA-DAI is a middleware which helps with data access and integration. The data can come from multiple sources through a computational grid. The functionality of this set of tools is oriented to the presentation of data resources as XML databases or relational databases. Various interfaces are offered and many relational database management systems (RDBMS) are supported. It also includes a set of components for query, transformation, and delivery of data in different ways and a simple toolkit to develop client applications. OGSA-DAI allows access via Web services to the XML or relational databases. The Web services allow query, transformation delivery, and update of the data. The OGSA-DAI Web services allow query, update, transformation, and data delivery offering data access integration services.
The following advantages were conclusive when the OGSA-DAI middleware for mantisGRID was selected: (1) Consistent and independent data and resource access. Each Data Source or node of mantisGRID belonging to an institution has its own uploading GRID portal or application. The application allows access and storage of structured reports containing persistent DICOM, XML, or other objects. (2) Grid data integration support through data resources. Data resources are databases available on the grid. In mantisGRID, each data resource is a node corresponding to each organization considered in the proposed architecture. (3) It provides important Web services for data federation and DQP (via OGSA-DQP version 3.2.1).
The DQP system allows the evaluation of queries generated over distributed data sources. These queries are processed through the grid via Web services provided by OGSA-DAI together with the Globus Toolkit. The framework based on DQP lays down the setting up of a coordinator and one or more evaluators whose interaction will render the functionality of the DQP.
The OGSA-DQP Coordinator is the main interaction point for all clients. It makes execution plans on distributed queries along multiple GRID nodes. The coordinator is currently implemented as a set of data resources and OGSA-DAI activities. Inside the implementation of the OGSA-DAI server, a node has been selected as coordinator of the abstraction and DICOM images request process on the grid.
Implementing the OGSA-DQP Coordinator implies to establish two application containers made by the Apache Tomcat Application Server (version 5.5). The main idea of the Apache Tomcat containers is to allow lodging and access for both OGSA-DAI and Globus Toolkit Web services within the architecture. Both containers must be instantiated at different ports in order to be enabled and the Globus Toolkit and must be deployed within Tomcat.
The Query Evaluation Service is used by the coordinator to execute its query plans. The existence of one or more evaluators is considered in the normal operation of OGSA-DQP, each evaluator carries out the queries assigned by the coordinator. Each evaluator has a XML document used to structure the queries gradually assigned by a coordinator. The structure is based on the DataResource ID or node where the request came from. A priority for each request is determined based on which node submitted the query first and the complexity of the query.
These queries are labeled within an execution plan and many of them can exist at a given time to be assigned to each evaluator (one to each evaluator). There is also the possibility that a query or query plan has to be distributed (depending on its complexity) among the evaluators available for its processing. When many queries exist, the partitioning of the plan containing the queries to be executed is crucial. These queries are executed on the distributed databases through mantisGRID. At this moment, each evaluator must handle a partition of such query plan for execution. In this way, the other evaluators will handle simultaneously the remaining partitions in order to approach the query execution plans generated by a coordinator within the grid.
Two evaluators are considered initially in the OGSA-DQP implementation for mantisGRID. These evaluators are considered necessary for the interaction in the DQP because the platform is oriented only toward DICOM medical images management and there are no levels of complexity that require the query partitioning process among evaluators.
Each node, which corresponds to an organization, has the following characteristics regarding the tools installed for mantisGRID:
One special node that contains a Globus Server (installation plus grid security infrastructure [GSI] with digital certificates and a digital certificate propagation service named MyProxy Server), the functionality of a OGSA-DAI server (DQP coordinator), the mantisGRID database, and the mantisGRID Web application that allows the interaction of the users with the grid.
The rest of the nodes contain the DQP evaluators, the corresponding mantisGRID databases and the compiled Globus Toolkit 4.2.0, the services of a simpleCA certifying entity, and a MyProxy client for the GRID certificates management.
Regarding the grid data integration, an OGSA-DAI service is used to accomplish this task. One node of the grid is the master node in charge of managing the data sources and guarantees the security at the database level. The software tools used to implement the OGSA-DAI service are: (1) the precompiled Globus Toolkit 4.0.5 from OGSA-DAI to deploy the Globus Web services via the Web Application Tomcat, (2) OGSA-DAI 3.0 to initially deploy the OGSA middleware in the Apache Tomcat containers, and (3) OGSA-DQP 3 package for the DQP.
DICOM and SR-CDA HL7 Interoperability in mantisGrid
Nowadays, great diversity of medical imaging devices in healthcare environments are DICOM compliant. The DICOM standard is a nonproprietary protocol for medical data exchange.21 It defines a digital image format and file structure for images and their related information.22 DICOM specifically describes: (1) information content, including structure and coding; (2) DICOM services for information management; and (3) messages protocol.23 It is widely used in diverse medical specialties like radiology, cardiology, and oncology, among others. From 1983 to date, DICOM has evolved into a robust standard applicable to a variety of medical systems, allowing handling medical images and derived structured documents as well as managing related workflow.24
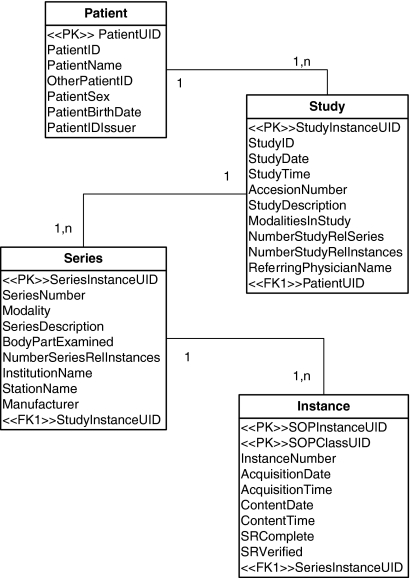
Unfortunately, middleware applications, such as the Globus Toolkit, lack the full integration of the DICOM standard25,26 since they are not targeted for a specific industry (e.g., Healthcare); therefore, this fact motivated us to work on its implementation and representation within the mantisGRID. For DICOM metadata management in mantisGRID, a relational database is used. Its design follows the outline of the information model of composite information object definition (IOD) from the DICOM standard27 and supplement 130;28 this model includes the patient, study, series, and instances objects; see Figure 2. Due to the semistructured information of diverse DICOM objects, there was a need to define “required-use” key fields as proposed in the DICOM information model Patient Root Query/Retrieve. Also, other fields were considered based on consultations to clinical experts to determine the best clinical practice advice. Figure 3 illustrates relational database tables defined for DICOM metadata management. This database schema includes a “Patient UID,” which consist in globally unique identifiers (UIDs). The UIDs are assigned by a coordinator organization in mantisGRID. This guarantees that a patient has a UID among the mantisGRID institutions. Each UID consists of a root and suffix component. Subcomponents are separated by a dot. The organizational root (org root) was assigned and registered by the Internet Assigned Numbers Authority (IANA). The suffix is internally created and subdivided in two subcomponents to guarantee internal uniqueness. The first one, <Patient ID code System> refers to the type of identifier, in this case will be the patient identifier. The second one, <Patient ID Seq> is the patient consecutive number. In summary, a patient UID will be of the form: Patient UID=<org root>.<Patient ID code System>.<Patient ID Seq>.
Fig 2.
DICOM composite instance IOD information model. Adapted from the DICOM Standard26 and supplement 130.27 This model includes the patient, study, series, and instances objects.
Fig 3.
Relational model for the DICOM header on the mantisGRID database.
The DICOM IODs currently supported by mantisGRID include the VL Photographic, Computed Tomography Image, Digital X-ray Image, Digital Intraoral X-ray Image, and Comprehensive SR. These IODs are represented using XML documents and XML schemas. This representation is accordance with work done in this area by diverse authors.22,29–33 The use of these languages allows detailed description of DICOM structures, along with their content, relationships, restrictions, standard lexicon, and their validation in conformity with the standard. In addition, its usability is optimal, entitling the splits of an XML schema document into several documents when it becomes large. This characteristic increases the readability, reusability, accessing control, and maintenance of XML schema documents.32 Technically, these XML schema documents are used in the creation and validation of XML documents to compile and represent clinical and technical information associated to IODs. Furthermore, reduces the ambiguity of reports in natural language and enhances the precision, clarity, and value of clinical documents.
Currently, the HL7 standard represents the foundation of many healthcare information management systems at the local and national levels. HL7 specifies structures and mechanisms to describe and communicate administrative and clinical data without focusing on a specific healthcare domain or communication technology.34 One of the most important domains in HL7 is the CDA. The CDA is a XML-based document markup standard that specifies the structure and semantics of clinical documents for the purpose of exchange. Thanks to the use of XML-based technologies and DICOM–HL7 standards committee results,35 constrained DICOM SR documents based on DICOM SR templates 2000 (Diagnostic Imaging Report Transformation Guide) and 7000 (General Relevant Patient Information) were mapped to HL7 CDA. This interoperability was achieved using XSLT tools such as Altova Mapforce® and the methodology proposed by Blazona et al.36
The acquisition of clinical and technical information in dermatological and dentistry DICOM studies was achieved by the implementation of a Web-feeding portal and standardized photography protocols. This Web application was developed using Java J2EE, Struts 1.2, DOM, and MyEclipse 7.0 M2 as integrated development environment. The validation of the DICOM standard, for the created studies, was accomplished with the “DICOM Validation Tool” from the Osirix Viewer. A sample screenshot of the Web interface created and a typical medical photography image are shown in Figure 4. OSirix is an image-processing open source software dedicated to DICOM images and is, at the same time, a DICOM PACS workstation for imaging and an image-processing software for medical research (radiology and nuclear imaging), functional imaging, 3D imaging, confocal microscopy, and molecular imaging.37
Fig 4.
Screenshots of the mantisGRID website and the DICOM viewer employed by the application.
Security
DICOM studies (imaging and reports) can be a source of relevant information for teaching and research activities. However, these studies contain information regarding patient's health condition and direct or indirect data about his/her identity.38–40 This confirms that patient medical records, in this case DICOM studies, are highly confidential sensitive documents. To guarantee this level of security, mantisGRID must fulfill the following requirements:
User authorization/authentication using mantisGRID certificates and database validation.
Interinstitution communication using the Secure Sockets Layer.
DICOM studies stores anonymized.
Based on the work performed by Suzuki et al.,38 Bland et al.,39 and legal rules from the Health Insurance Portability and Accountability Act40,41 and the Colombian government,42 the desired de-identification features of mantisGRID were identified. The anonymizer application removes identifiers from the patient metadata at each institution's database according to privacy policies defined by the each institution/node. Anonymization will be of value for research purposes, such as in epidemiological studies. As a result, some users of the grid would only be able to access anonymized data. Alternatively, when the mantisGrid platform is used for clinical purposes, patient identifiers would be available, given that the user has the required privileges. In either case, data security is carefully treated to maintain patient confidentiality.
Authentication/Authorization
Authentication is performed through certificates, using the GSI tools from Globus Toolkit. In this case, we used the SimpleCA (Certificate Authority) tool, allowing confidence degrees between grid nodes and the CA server. Specifically, we use MyProxy Server middleware43 regarding authentication with CA Server, propagation into mantisGRID, and Time to Lives assignation.
The authorization is made on RDBMS using the USER-ROL model, which allows the user to set different actions. One advantage of this approach is that mantisGRID can be adapted for exchanging security attributes with other grid networks through shibboleth middleware.44
Performance TEST
The following is a description of the experiments developed to test the performance of mantisGRID when working with heterogeneous environments over the RENATA network (currently 30 Mbps but with very low traffic). The experiments were conducted over a variety of machines with different database across different domains.
In order to guarantee the privacy of the data, all the medical and radiological information stored in the system was anonymized after acquisition as explained before. The databases were located at the three main participant institutions and interconnected via the RENATA network. Even though the system supports heterogeneous database engines, all of them were developed using mySQL in order to improve the efficiency and reduce the administration task of the system.
Two different experiments were designed in order to test the performance of the system. The first one with the aim to test the grid database environment, and the second one with the purpose of testing file transfer capabilities across the mantisGRID nodes.
Database Engine Test
The database engine tests consisted in a series of queries to the table with higher complexity available in the database system. Each query was run multiple times and at different times of the day, this way the network inconsistencies could be isolated providing a mean value. The queries designed for the test included only the medical information and metadata from the DICOM files. There was no image transfer during this test. As all the databases at the nodes used mySQL, it was possible to compare the performance of mantisGRID (OGSA-DAI) against the Distributed mySQL scheme. A third scenario considered only local queries so there was no network delay.
The results are shown in Table 1 and plotted in Figure 5. The first column contains the number of records accessed by the database during the test, the other columns contains the mean time in seconds employed by the three different schemes and its standard deviation (SD). The minimum time in all the cases was obtained by the local queries as expected. This value was kept only as a reference as it does not apply to a grid environment. It can be observed that the mantisGRID performance is below that achieved by the Distributed mySQL scheme. This can be explained by the additional processing required by the OGSA-DAI model. Firstly, OGSA-DAI uses Web services (evaluator and coordinator process) to cope with heterogeneous systems. Secondly, as OGSA-DAI works natively with Java while the mantisGRID was developed under PHP, it was necessary to translate the query results from Java into PHP code. This translation also introduces a performance overhead in mantisGRID. However, in spite of the additional processing, the performance seems to improve when the amount of records fetched increases. Notice that, as the problem scales, the performance difference between OGSA-DAI and Distributed mySQL decreases up to 10% and then the difference is maintained. This behavior is observed in Figure 5 where, after 5,000 records, the two plots have approximately the same slope.
Table 1.
Database Engine Mean Access Time and its SD (in Seconds)
| Records | MySQL | SD (MySQL) | Distributed mySQL | SD (Distrib MySQL) | mantisGRID (OGSA-DAI) | SD (mantisGRID) |
|---|---|---|---|---|---|---|
| 8 | 0.00 | 0.00 | 0.18 | 0.03 | 4.03 | 0.60 |
| 800 | 0.00 | 0.00 | 2.30 | 0.35 | 4.76 | 0.71 |
| 1,500 | 0.02 | 0.00 | 5.60 | 0.84 | 6.10 | 0.92 |
| 5,000 | 0.19 | 0.02 | 7.45 | 0.75 | 8.20 | 0.82 |
| 10,000 | 0.36 | 0.04 | 8.30 | 1.25 | 9.10 | 1.37 |
| 15,000 | 0.44 | 0.04 | 9.00 | 1.85 | 9.90 | 1.98 |
Fig 5.
Plot of the database engine access time versus the number of records fetched. The MySQL time corresponds to local access time (no network involved) and is included as reference only. The overhead of mantisGRID stabilizes as the number of records increases. mantisGRID allows queries to heterogeneous databases engines.
File Trower Test
The File Trower is a simple file-moving application that allows to transfer files from one server to another in a secure way, all this under the premise that “a study” should always be in its home server and should only be seen (moved) by others upon request. The decision to create this application was made after analyzing various alternatives like the Network File System (NFS) and HTTP-to-HTTP. NFS is not robust enough for our purposes and sharing is out of the question due to privacy requirements. HTTP-to-HTTP added complexity to the systems since security of the Web server had to be addressed by each institution and a simple application for moving the file had to be written. The File Trower was designed to fulfill all these necessities in a simple way.
The mantisGRID operates on a variety of digital objects as shown in Table 2. The size of the objects varies depending on the modality of the exam: intraoral photography, dental panoramic X-ray, orthodontic cephalogram, dental X-ray bitewing, skin lesion photography, CT of the head, and an MRI scan of the brain. From these, the maximum file size corresponds to 22 MB for the skin lesion photography case. The File Trower application was designed to move these files across the net. To assess its performance, a simple test consisting of moving files across the grid and comparing the results with the HTTP mechanism was accomplished. The results are summarized in Table 3. Two hundred files where transmitted in each case at different times of the day. The results presented in the table correspond to the average value. The file transfer was accomplished with files up to 100 MB in size. It can be observed that, for files in the order of 10 to 50 MB, the performance of File Trower is comparable to a direct download from a HTTP server (Apache 2). For higher file sizes, the performance of HTTP is better than File Trower. However, as seen before, the maximum file size that is transmitted via mantisGRID is of 22 MB.
Table 2.
Average Size of the Medical Data and its SD
| Modality | Type | Width (pixels) | Height (pixels) | Size (MB) | SD (MB) |
|---|---|---|---|---|---|
| XC | Intraoral photography | 916 | 600 | 1.64 | 0.18 |
| PX | Dental panoramic X-ray | 1,339 | 700 | 0.85 | 0.05 |
| RG | Orthodontic cephalogram | 1,200 | 1,060 | 1.20 | 0.13 |
| IO | Dental X-ray bitewing | 956 | 388 | 0.37 | 0.17 |
| XC | Skin lesion photography | 2,477 | 3,018 | 22.17 | 5.71 |
| CT | CT of the head | 512 | 512 | 0.60 | 0 |
| MR | MR of the brain | 256 | 256 | 0.40 | 0 |
Table 3.
File Transfer Comparison Between File Trower and HTTP Methods (Time and SD in Seconds)
| Size (MB) | File Trower | SD | HTTP (Apache 2) | SD |
|---|---|---|---|---|
| 20 | 17 | 2.31 | 20 | 6.57 |
| 50 | 51 | 3.12 | 48 | 4.28 |
| 100 | 105 | 5.11 | 99 | 9.04 |
Discussion and Future Work
The mantisGRID project aims to provide Grid services for Colombia and Latin America. In its first phase of development, a reliable architecture has been created by the use of OGSA-DAI services and the Globus Toolkit in addition to a PHP-based website to run the Web interface. Special attention was made to make the mantisGRID fully compliant with both DICOM and the HL7 standards, as well as to guarantee data security and integrity while always maintaining patient data confidentiality. Data transfer across the current nodes of the grid was securely accomplished via an application called File Trower and employed the RENATA network. Computational experiments demonstrated that appropriate transfer rates are achievable especially for a large number of records. The results obtained are comparable to those achieved by others.45
Arguably, usage is the most important task for a healthgrid following its implementation and there are indeed few reports of both successful implementation and, more importantly, evaluation of healthgrids clinical outcomes.46 To address this challenge, the mantisGRID has started to work with few nodes and across institutions located in Colombia to allow ongoing and flexible collaborations during the developing phase of the grid. Furthermore, in a second phase of the mantisGRID project, we are fully focusing on two applications: medical digital photography for dermatology and dentistry. At the same time, the mantisGRID team is working to attract institutions to be added as nodes and collaborators in Colombia and also seeking to expand to other regions in Latin American.
The mantisGRID team has also identified that standards and protocols are of foremost importance to guarantee successful deployment of the healthgrids. In addition to complying with both DICOM and HL7,47 we plan on adopting initiatives of controlled vocabularies extending for example to image annotation and markup, as recently proposed by the caBIG team.48 Furthermore, we are working intensively on protocols for medical light photography where lighting conditions and color preservation are crucial to enable distant collaboration or telemedicine.16
Healthgrids initiatives, almost by definition, require work in collaboration and we are keen to discuss with potential partners and users how to shape the mantisGRID to provide effective healthgrid services for Colombia and the Latin American communities.
Acknowledgement
The authors would like to express their thanks to the sponsoring institutions Universidad EAFIT, Universidad CES, Clinica CES, and Escuela de Ingenieria de Antioquia, as well as the Biomedical Imaging Resource (BIR) from Mayo Clinic. This work was cofunded by “Ministerio de Educación Nacional” Colombia, Red Nacional Académica de Tecnología Avanzada (RENATA).
Contributor Information
Manuel Garcia Ruiz, Phone: +57-4-2619500, FAX: +57-4-2664284, Email: mgarcia@eafit.edu.co.
Alvin Garcia Chaves, Email: agarcia@ces.edu.co.
Carlos Ruiz Ibañez, Email: cruiz@eia.edu.co.
Jorge Mario Gutierrez Mazo, Email: vadersolo@gmail.com.
Juan Carlos Ramirez Giraldo, Email: RamirezGiraldo.JuanCarlos@mayo.edu.
Alejandro Pelaez Echavarria, Email: apelaez@ces.edu.co.
Edison Valencia Diaz, Email: pfevalencia@eia.edu.co.
Gustavo Pelaez Restrepo, Email: bmguale@eia.edu.co.
Edwin Nelson Montoya Munera, Email: emontoya@eafit.edu.co.
Bernardo Garcia Loaiza, Email: bgarcial@eafit.edu.co.
Sebastian Gomez Gonzalez, Email: sgomezg2@eafit.edu.co.
References
- 1.Diarena M, Nowak S, Boire JY, Bloch V, Donnarieix D, Fessy A, Grenier B, Irrthum B, Legre Y, Maigne L, Salzemann J, Thiam C, Spalinger N, Verhaeghe N, Vlieger P, Breton V. HOPE, an open platform for medical data management on the grid. Stud Health Technol Inform. 2008;138:34–48. [PubMed] [Google Scholar]
- 2.Breton V, Dean K, Solomonides T: The Healthgrid White paper. Proceedings of Healthgrid Conference, IOS Press, 112, 2005 [PubMed]
- 3.Jacq F, Bacin F, Meda N, Donnarieix D, Salzemann J, Vayssiere V, Renaud M, Traore F, Meda G, Nikiema R, Jacq N, Breton V: Towards grid-enabled telemedicine in Africa. Proceedings of IST-Africa 2006 Conference, Pretoria, May 2006
- 4.Amendolia SR, Estrella F, Hassan W, Hauer T, Manset D, McClatchey R, Rogulin D, Solomonides T. MammoGrid: a service oriented architecture based medical grid application. In: Jin H, Pan Y, Xiao N, Sun J, editors. Lecture Notes in Computer Science, GCC 2004, LNCS 3251. Berlin: Springer; 2004. pp. 939–942. [Google Scholar]
- 5.Montagnat J, Breton V, Magnin IE. Partitioning medical image databases for content based queries on a grid. Methods Inf Med. 2005;44(2):154–160. [PubMed] [Google Scholar]
- 6.Sharma A, Pan T, Barla Cambazoglu B, Gurcan M, Kurc T, Saltz J. VirtualPACS—a federating gateway to access remote image data resources over the grid. J Digit Imaging. 2009;22(1):1–10. doi: 10.1007/s10278-007-9074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics. 2006;22:1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]
- 8.Freund J, Ioannis Y, Liu P, McClatchey R, Morley-Fletcher E, Pennec X, Pongiglione G, Zhou X. Health-e-Child: an integrated biomedical platform for grid-based paediatrics. Stud Health Technol Inform. 2006;120:259–270. [PubMed] [Google Scholar]
- 9.Pernod E, Souplet JC, Rojas Balderrama J, Lingrand D, Pennec X: Multiple sclerosis brain MRI segmentation workflow deployment on the EGEE Grid. Proceedings of the MICCAI-Grid Workshop, New York, Sept 2008
- 10.Bonneto P, Oliva G, Formiconi AE: A medical imaging environment based on a grid computing infrastructure. Proceedings of the 25th Annual International Conference of the IEEE EMBS, Cancun, Mexico, Sept 2003
- 11.Johnson TR, Krauss B, Sedlmair M, Grasruck M, Bruder H, Morhard D, Fink C, Weckbach S, Lenhard M, Schmidt B, Flohr T, Reiser MF, Becker CR. Material differentiation by dual energy CT: initial experience. Eur Radiol. 2007;17(6):1510–1517. doi: 10.1007/s00330-006-0517-6. [DOI] [PubMed] [Google Scholar]
- 12.Jacq N, Legré Y, Medernach E, Montagnat J, Maaß A, Reichstadt M, Schwichtenberg H, Sridhar M, Kasam V, Zimmermann M, Hofmann M, Breton V. Grid-enabled virtual screening against malaria. J Grid Comput. 2008;6:29–43. doi: 10.1007/s10723-007-9085-5. [DOI] [Google Scholar]
- 13.Jacq F, Bacin F, Meda N, Donnarieix D, Salzemann J, Vayssiere V, Jacq N, Renaud M, Traore F, Meda G, Nikiema R, Breton V: Towards grid-enabled telemedicine in Africa. Proceedings of the IST-Africa 2006 Conference Information Society Technologies in Africa, Prétoria, South Africa, 2006
- 14.Marechal B: Grid infrastructure for e-Science: a use case from Latin America and Europe. Proceedings of EuroAfriCa-ICT FP7 Awareness Workshop, Kampala, Uganda, Oct 2008
- 15.García A, Ruiz C, Valencia E, Ramirez JC, Pelaez A, Montoya E, et al: Modelación de la plataforma Mantis-GRID para la gestión e integración de repositorios distribuidos de imágenes médicas. Memorias III Congreso Colombiano de Bioingeniería e Ingeniería Biomédica [CD-ROM], June 4–6, Pereira, Colombia, 2008
- 16.García A, Ruiz C, Valencia E, Ramirez JC, Pelaez A, Montoya E, et al: mantisGRID: a grid platform for DICOM medical images management. Pan American Health Care Exchanges Proceedings, [CD-ROM], March 16–20, Mexico City, Mexico, 2009
- 17.Grant A, Antonioletti M, Hume AC, Krause A, Dobrzelecki B, Jackson MJ, Parsons M, Atkinson MP, Theocharopoulos E: OGSA-DAI: middleware for data integration: selected applications. eScience, IEEE Fourth International Conference on eScience ’08, pp. 343–343, 7–12 Dec. 2008
- 18.OGSA-DAI: The OGSA-DAI Project. Available at: http://www.ogsadai.org.uk/. Accessed April 2009
- 19.Foster I. Globus Toolkit version 4: software for service-oriented systems. J Comput Sci Technol. 2006;21(4):513–520. doi: 10.1007/s11390-006-0513-y. [DOI] [Google Scholar]
- 20.The Globus Alliance. Globus Toolkit. Available at: http://www.globus.org. Accessed April 2009
- 21.Part 1: introduction and overview. Rosslyn: National Electrical Manufacturers Association; 2008. [Google Scholar]
- 22.Tirado-Ramos A, Hu J, Lee KP. IOD-based UML representation of DICOM structured reporting: a case study on transcoding DICOM to XML. J Am Med Inform Assoc. 2002;9:63–71. doi: 10.1136/jamia.2002.0090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chartier A. DICOM (Digital Imaging and Communications in Medicine) in dermatology. In: Wooton R, Oakley A, editors. Teledermatology. UK: The Royal Society of Medicine Press Limited; 2002. p. 205. [Google Scholar]
- 24.Digital Imaging and Communications in Medicine (DICOM). DICOM Brochure. Rosslyn, VA: National Electrical Manufacturers Association, 2008. Available at: http://medical.nema.org. Accessed April 2009
- 25.Erberich SG, Silverstein JC, Chervenak A, Schuler R, Nelson MD, Kesselman C. Globus MEDICUS-Federation of DICOM medical imaging devices into healthcare grids. Stud Health Technol Inform. 2007;126:269–278. [PubMed] [Google Scholar]
- 26.Vossberg M, Tolxforff T, Krefting D. DICOM Image communication in Globus-based medical grids. IEEE Trans Inf Technol Biomed. 2008;12(2):145–153. doi: 10.1109/TITB.2007.905862. [DOI] [PubMed] [Google Scholar]
- 27.National Electrical Manufacturers Association (NEMA): Digital imaging and communications in medicine (DICOM), part 3: information object definitions. Rosslyn, VA, 2008. Available at: http://medical.nema.org/
- 28.National Electrical Manufacturers Association (NEMA): Digital imaging and communications in medicine (DICOM), supplement 130: ophthalmic refractive measurements storage and SR SOP classes, NEMA, Rosslyn, VA, Version: Final Text 24 January, 2008. Available at: http://medical.nema.org/
- 29.Noumeir R. DICOM structured report document type definition. IEEE Trans Inf Technol Biomed. 2003;7(4):318–328. doi: 10.1109/TITB.2003.821334. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Lee KP, Hu J. Generating XML schemas for DICOM structured reporting templates. J Am Med Inform Assoc. 2005;12(1):72–83. doi: 10.1197/jamia.M1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KP, et al. Expressing DICOM SR constraints in XML. In: Lemke HU, et al., editors. Proceedings of CARS 2002. New York: Springer; 2002. pp. 491–496. [Google Scholar]
- 32.Lee KP, Hu J. XML schema representation of DICOM structured reporting. J Am Med Inform Assoc. 2003;10:213–223. doi: 10.1197/jamia.M1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J, Lee KP: DICOM XML DTD/Schema Generator. World Intellectual Property Organization, 2001
- 34.HL7 Inc: Official Web Page. Available at: http://www.hl7.org. Accessed January 2009
- 35.DICOM Standards Committee: Supplement 135: SR Diagnostic Imaging Report Transformation Guide. Draft 04 for DICOM Public Comment and HL7 1st Informative Ballot. Available at: http://www.hl7.org. Accessed January 2009
- 36.Blazona B, Koncar M. HL7 and DICOM based integration of radiology departments with healthcare enterprise information systems. Int J Med Inform. 2007;76(Suppl 3):425–432. doi: 10.1016/j.ijmedinf.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Osirix Imaging Software: Available at: http://www.osirix-viewer.com/index.html. Accessed January 2009
- 38.Suzuki H, Amano M, Kubo M, Kawata Y, Niki N, Nishitani H: Anonymization server system for DICOM images. Proc SPIE, 2007. doi:10.1117/12.709947
- 39.Bland PH, Laderach GE, Meyer CR. A Web-based interface for communication of data between the clinical and research environments without revealing identifying information. Acad Radiol. 2007;14(6):757–764. doi: 10.1016/j.acra.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fetzer T, West OC. The HIPAA privacy rule and protected health information: implications in research involving DICOM image databases. Acad Radiol. 2007;15(3):390–395. doi: 10.1016/j.acra.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Office for Civil Rights–HIPAA [WebSite]*.Washington: U.S. Department of Health & Human Services. Available at: http://www.hhs.gov/ocr/hipaa/. Accessed January 2009
- 42.Ministerio de la Protección Social. Resolución 1995 del 8 de Julio de 1999. Bogota: Ministerio de Salud, 1999. Available at: http://www.minproteccion-social.gov.co. Accessed January 2009
- 43.MyProxy Credential Management Service: Available at: http://grid.ncsa.uiuc-.edu/myproxy. Accessed April 2009
- 44.The Globus Alliance: Grid Ecosystem-Shibboleth. Available at: http://www.-globus.org/grid_software/security/shibboleth.php. Accessed April 2009
- 45.Luo Y, Jiang L, Zhuang T. A grid-based model for integration of distributed medical databases. J Digit Imaging. 2008;22:579–588. doi: 10.1007/s10278-007-9077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estrella F, Hauer T, McClatchey R, Odeh M, Rogulin D, Solomonides T. Experiences of engineering Grid-based medical software. Int J Med Inform. 2007;76:621–632. doi: 10.1016/j.ijmedinf.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 47.García A, Ruiz C, Valencia E, Ramirez JC, Pelaez A, Montoya E, et al: Representación de objetos DICOM en XML para uso en repositorios distribuidos. Memorias III Congreso Colombiano de Bioingeniería e Ingeniería Biomédica [CD-ROM],Junio 4–6; Pereira, Colombia, 2008
- 48.Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL: The caBIG annotation and image markup project. J Digit Imaging 2009 doi:10.1007/s10278-009-9193-9 [DOI] [PMC free article] [PubMed]