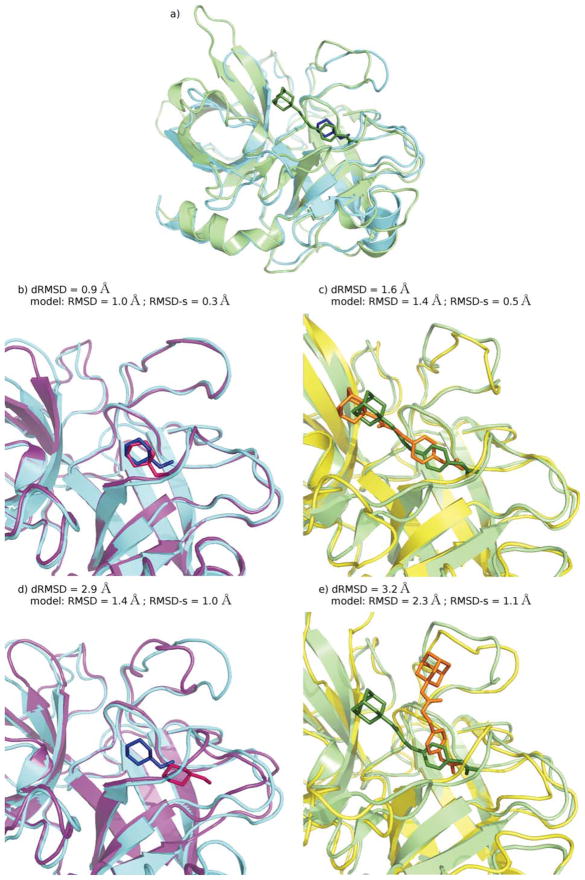
Figure 9.
Some docking results for the 1EJN + 1TNG subset of protein–ligand complexes: Urokinase-type plasminogen activator - N-(1-adamantyl)-N′-(4-guanidinobenzyl) urea complex + Trypsin - Aminomethylcyclohexane complex. a) The two experimental structures of the complexes upon structural superimposition of the proteins: 1EJN, protein and ligand are colored in green; 1TNG, protein in cyan, ligand in blue; b), d) 1TNG: two binding geometries obtained by docking calculations for protein models of different qualities (models are colored in purple and ligands in pink), compared to the experimental geometry (cyan and blue); c), e) 1EJN: two binding geometries obtained by docking calculations for protein models of different qualities (yellow and orange), compared to the experimental geometry (green).