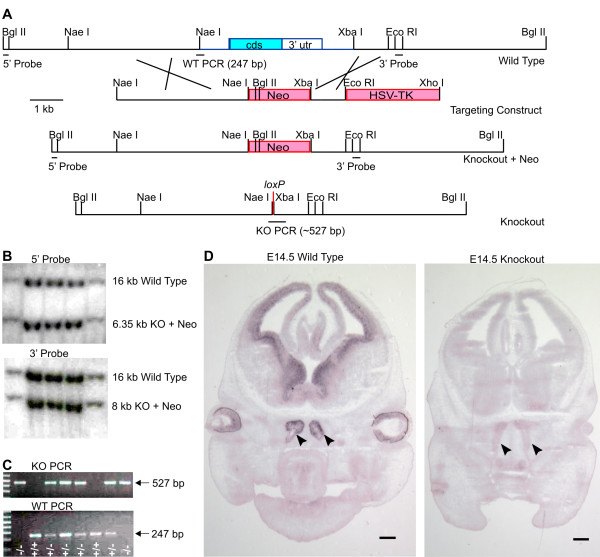
Figure 1.
Generation of Insm1 knockout mice. (A) Schematic of the recombination strategy used to generate the Insm1 knockout. Using homologous recombination in embryonic stem (ES) cells, we replaced a genomic DNA fragment of 3.4 kb (in blue/cyan) containing the entire single exon of Insm1 plus 927 bp of upstream and 955 bp of downstream sequence with a 'floxed' neomycin cassette (in red/pink). After generating chimeric mice and, from these, heterozygotes, the neomycin cassette was removed by crossing with E2a-Cre transgenic mice. Probes (5' and 3') used for Southern blot genotyping and DNA fragments obtained by PCR genotyping of the wild type, knockout + neomycin and knockout alleles are depicted. (B) Southern blots of BglII digests of genomic DNA from recombined ES cells with 5' and 3' probes reveal the expected bands from the knockout (KO) and wild type alleles and confirm that recombination was complete. (C) Genotyping PCR using genomic DNA obtained from a litter of embryos (embryonic day 10.5) resulting from crossing two heterozygous parents. Primers used are described in methods and their products depicted in (A). The litter contained two knockouts (-/-), two wild types (+/+) and four heterozygotes (+/-), as indicated in the figure. (D) In situ hybridization (ISH) with an antisense probe against the 3' UTR of Insm1 on wild type and knockout littermate embryos (E14.5, coronal sections) reveal the presence of Insm1 mRNA in the brain, retina and olfactory epithelium (arrowheads) of wild type but not of knockout embryos, functionally confirming the knockout of Insm1. Scale bars: 500 μm.