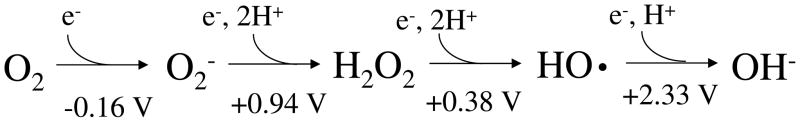Abstract
Life evolved in an anaerobic world; therefore, fundamental enzymatic mechanisms and biochemical pathways were refined and integrated into metabolism in the absence of any selective pressure to avoid reactivity with oxygen. After photosystem 2 appeared, environmental oxygen levels rose very slowly. During this time microorganisms acquired oxygen tolerance by jettisoning enzymes that use glycyl radicals and low-potential iron-sulfur clusters, which can be directly poisoned by oxygen. They also developed mechanisms to defend themselves against O2− and hydrogen peroxide, partially reduced oxygen species that are generated as inadvertent by-products of aerobic metabolism. These species are more chemically reactive than is molecular oxygen itself. Contemporary organisms have inherited both the vulnerabilities and the defenses of these ancestral microbes. Current research seeks to identify these, and bacteria comprise an exceptionally accessible experimental system that has provided the many of the answers. This manuscript reviews recent developments and identifies remaining puzzles.
Keywords: OxyR, PerR, SoxR, Fenton reaction, iron, hydroxyl radical
Introduction
Virtually all organisms maintain high titers of enzymes that scavenge superoxide (O2−) and hydrogen peroxide (H2O2). This fact suggests the hypothesis that has driven research in oxidative stress for the past thirty years (1): that oxygen toxicity is primarily mediated by partially reduced oxygen species that are more reactive than is molecular oxygen itself (Fig. 1). Such species are inevitable by-products of aerobic metabolism, and the evolution of enzymes that scavenge them was an adaptation that allowed ancient microbes to occupy aerobic habitats.
Fig. 1.
The redox states of oxygen with standard reduction potentials. The standard concentration of oxygen was regarded as 1 M.
Current research attempts to identify the sources of these oxidants and the biomolecules that they damage. In addition, it seeks to discover additional protective strategies that complement the action of scavenging enzymes. These defenses evolved in the pre-eukaryotic world and are broadly conserved among diverse life forms. Work has proceeded in a variety of model organisms; however, bacteria are unequivocally the experimental systems in which progress has been most rapid.
There are great advantages in using bacteria to illuminate the processes of oxidative stress; these have had such an impact that they are worth enumerating here. First, workers have been able to knock out multiple scavenging enzymes and thereby reveal the consequences of chronic O2− and H2O2 stresses. Second, because growth conditions can be manipulated to isolate catabolic and biosynthetic pathways, experimenters have been able to identify the processes and biomolecules that are most vulnerable to oxidants. Third, the absence of organelles allows one to measure or calculate concentrations of metabolites and oxidants and thus to appraise whether chemical reactions that are observed in vitro are likely to occur at significant rates in vivo. Fourth, the ability of E. coli to grow anaerobically has allowed workers to construct mutants that lack key oxidative defenses—and to observe the impact when oxygen is subsequently introduced. And finally, because microbes have little control over their extracellular environments, they acquired mechanisms that sharply adjust the synthesis of defensive proteins in response to stress. Investigators have used genetic and genomic methods to dissect these circuits, thereby pinpointing genes that play important roles in protecting cells from stress. The purpose of this review is to summarize what has been learned and to emphasize some of the important mysteries that remain.
1. Mechanisms of superoxide and hydrogen peroxide toxicity
1.1. The formation of reactive oxygen species
Oxygen crosses membranes so freely (2) that the intracellular concentration is essentially equivalent to that which is immediately outside the cell. Partially reduced oxygen species are generated when molecular oxygen adventitiously abstracts electrons from the exposed redox moieties of electron-transfer enzymes. Flavoenzymes in particular have been identified as culprits (3), and since this class of enzyme is ubiquitous and abundant, it follows that all aerobic organisms experience a steady flux of endogenously generated oxidants. A mixture of O2− and H2O2 is formed, reflecting the fact that either one or two electrons can be transferred in an oxidation event (4). The overall reaction rate is proportional to collision frequency; thus, O2− and H2O2 fluxes depend directly upon the ambient concentration of oxygen. For this reason microaerophilic bacteria—and mammalian cells—are substantially protected from oxidative stress because they dwell in habitats where extracellular fluids are not fully saturated with air.
Hydrogen peroxide formation in E. coli has been directly measured by the rate at which H2O2 effluxes from strains that lack catalases and peroxidases (5). Approximately 15 μM/s H2O2 is formed in well-fed cells. The rate of O2− production has been estimated to be about 5 μM/s. Interestingly, in E. coli the predominant sources of cytoplasmic H2O2 must lie outside the respiratory chain, as the overall rate of H2O2 formation was not substantially diminished by mutations that eliminated respiratory enzymes. However, the respiratory chain was the major source of O2− that was released into the periplasm on the external face of the cytoplasmic membrane (6).
Basal oxidative defenses are sufficient to protect bacteria from the O2− and H2O2 that are formed by enzyme autoxidation. However, most microbes induce additional responses when elevated levels of O2− and H2O2 stress are artificially imposed in the laboratory. This raises the question: What are the natural sources of oxidative stress that selected for the evolution of these extra defenses?
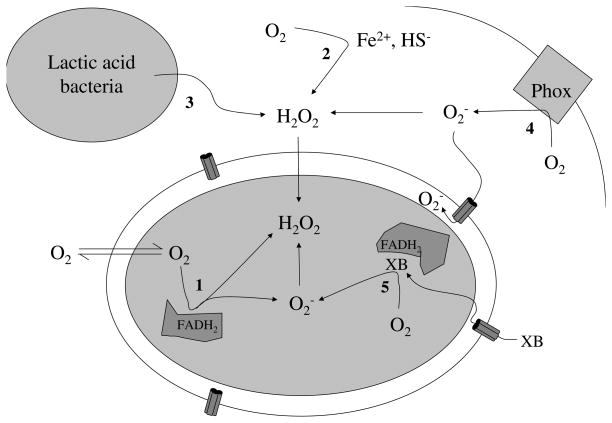
Several sources have been identified (Fig. 2). The natural vulnerability of organisms to reactive oxygen species (ROS) has been targeted by plants and microbes that wish to suppress the growth of their competitors. They excrete redox-cycling compounds that diffuse into nearby bacteria, where the agents generate O2− by oxidizing redox enzymes and transferring the electrons to molecular oxygen. Such compounds can elevate the rate of intracellular ROS formation by orders of magnitude. They are potent inducers of the SoxR(S) regulon (7, 8), which commands the induction of a battery of defensive proteins, including superoxide dismutase.
Fig. 2.
Sources of oxidative stress for bacteria include (1) intracellular enzyme autoxidation, (2) environmental redox reactions, (3) H2O2 released by competing microbes, (4) phagosomal NADPH oxidase, and (5) redox-cycling antibiotics.
Hydrogen peroxide (unlike O2−) is an uncharged species that penetrates membranes; therefore, H2O2 stress arises inside cells whenever H2O2 is present in their extracellular environment. H2O2 can be formed by chemical processes when reduced metals and sulfur species seep from anaerobic sediments into oxygenated surface waters. H2O2 is also produced through photochemical mechanisms; such processes can generate 1–20 micromolar H2O2 in sterile media that stands on the bench under room lighting, which can be an unrecognized source of oxidative stress in lab experiments. Such sources of H2O2 probably drove the evolution of the OxyR, PerR, and Yap-1 regulons, each of which induces H2O2 scavengers and other defensive enzymes in microbes.
These systems also defend microbes against H2O2 assault by their competitors. Redox-cycling drugs generate H2O2 in concert with O2−, due to dismutation of the latter. Lactic-acid bacteria suppress the growth of competing microbes by using pyruvate and lactate oxidases to excrete large doses of H2O2. More famously, H2O2 is actively generated by the NADPH oxidases that are activated along plant wound sites and by mammalian macrophages. Mammals that lack the latter enzyme are prone to persistent microbial infections.
1.2. What biomolecules do superoxide and H2O2 damage?
Despite the ubiquity of SOD and catalases, it proved difficult to demonstrate that O2− and H2O2 are important biological toxins. O2− did not damage biomolecules that were surveyed in vitro (9–11), a result that initially appeared to cast doubt on both its potency and the biological role of SOD. And while cells could be killed by exogenous H2O2, these experiments often required millimolar doses that were far beyond physiological relevance.
Clarity emerged from the construction of bacterial mutants that lacked scavenging enzymes. E. coli strains devoid of cytoplasmic SODs were constructed by Carlioz and Touati in 1986 (12). These strains grew well anaerobically but exhibited a variety of aerobic growth defects that derived from the accumulation of endogenous O2−. Similarly, E. coli catalase/peroxidase mutants are poisoned by the micromolar doses of intracellular H2O2 that accumulate (13, 14). A key goal has been to track these defects back to the biochemical lesions that cause them.
Both sets of mutants exhibit catabolic and biosynthetic defects that stem from the inactivation of a family of dehydratases (15, 16). These enzymes each contain a solvent-exposed [4Fe-4S]2+ cluster, including an under-coordinated iron atom that binds substrate and then abstracts a hydroxyl anion from it. Superoxide and H2O2 penetrate into the active site of these enzymes, bind the critical iron atom, and oxidize the cluster to a [4Fe-4S]3+ form that is unstable. The O2− univalently oxidizes the cluster; the [4Fe-4S]3+ product then releases the catalytic iron atom and is left in an inactive [3Fe-4S]+ form. The H2O2-oxidized cluster is divalently oxidized, probably in a two-step process, and releases ferric iron to generate the [3Fe-4S]+ species (14). The rate constants for the reactions by O2− (up to 106 M−1 s−1) and H2O2 (104 M−1 s−1) are high enough to suggest that dehydratase damage occurs continuously even in wild-type aerobic cells. Activity is preserved only because of processes that reactivate the enzymes (below).
Both O2− and H2O2 are also mutagenic (17, 18). H2O2 directly oxidizes unincorporated intracellular ferrous iron, some of which is associated with DNA (19, 20):
| (1) |
| (2) |
The rate constant of the Fenton reaction (eq. 1) depends upon the coordination environment of the iron atom. Early measurements conducted at acid pH indicated the value was quite low (21), but at physiological pH it has been measured to range from 5000 – 20,000 M−1 s−1 (13, 22). The hydroxyl radical that is generated reacts at nearly diffusion-limited rates near the site of its formation. In E. coli less than 1 micromolar of intracellular H2O2 is sufficient to cause crippling levels of DNA damage. It was initially suspected that the mutagenicity of O2− derived from its ability to recycle oxidized iron to the ferrous form; however, the intracellular concentration of O2− is too low for it to be a significant player relative to other metabolites, such as cysteine (23). Instead, O2− promotes DNA damage indirectly by releasing iron from damaged dehydratase clusters (24, 25). Since iron binds adventitiously to the surfaces of lipids and proteins, too, it seems probable that they also suffer some level of oxidation.
Other phenotypic defects of scavenger mutants have been identified but not yet explained on a biochemical level. Both SOD and catalase/peroxidase mutants of E. coli are incapable of synthesizing aromatic products, including amino acids. This deficiency may arise from the oxidation of the dihydroxyethyl thiamine intermediate of transketolase (26), but firm structural evidence is lacking. SOD mutants of both E. coli and S. cerevisiae are also unable to perform normal sulfur metabolism (12, 27); in this case the causal lesion remains completely unknown. It is likely that additional injuries remain to be identified.
2. Inducible responses to reactive oxygen species
2.1. The SoxR(S) regulator of the response to superoxide stress
In the mid-70’s Hassan and Fridovich discovered that manganese-containing superoxide dismutase (MnSOD) is strongly induced when E. coli is exposed to redox-cycling antibiotics (28). Ten years later, the Demple and Weiss labs independently determined that the response is governed by two proteins: SoxR, which is a sensor protein that detects redox stress, and SoxS, a transcriptional activator that positively regulates about two dozen genes around the chromosome (7, 8, 29) (Table 1). The resting SoxR protein is a homodimer that coordinates one [2Fe-2S]+ cluster per subunit. When redox-cycling agents were added to cells in which SoxR was overproduced, EPR spectroscopy detected the oxidation of the cluster to a +2 state (30, 31). In vitro inspection showed that this oxidation event caused a conformational change in the dimeric protein, perhaps due to the gain of electrostatic repulsion between the two oxidized clusters. This contortion is transmitted to the bound promoter region, thereby improving an RNA polymerase binding site that is otherwise disabled due to an overly short distance between −35 and −10 promoter elements (32).
Table 1.
Selected genes induced by the SoxRS systema.
| Oxidant-resistant dehydratase isozymes. | |
| fumC | Fumarase C. |
| acnA | Aconitase A. |
| Suspected cluster repair. | |
| yggX | Fe/S cluster repair protein? |
| zwf | Glucose-6-phosphate dehydrogenase |
| fpr | NADPH:flavodoxin/ferredoxin oxidoreductase |
| fldA | Flavodoxin A |
| fldB | Flavodoxin B |
| Drug efflux and/or resistance. | |
| acrAB | Drug efflux pump |
| tolC | OMP component of drug efflux pump |
| micF | OmpF antisense sRNA |
| marAB | Multiple antibiotic resistance operon |
| nfnB | Nitroreductase |
| rimK | Modification of ribosomal protein S6 |
| Other | |
| nfo | Endonuclease IV |
| fur | Iron-uptake regulatory protein |
| sodA | Manganese-containing superoxide dismutase |
| ribA | cGMP hydrolase |
Sources: [Pomposiello, 2001 #2922] and references within.
In enteric bacteria this process stimulates transcription of soxS by more than 20-fold. The SoxS protein, in turn, is a secondary transcription factor that enhances expression of genes listed in Table 1. In other bacteria, SoxR is the direct inducer of each member of the regulon (33, 34).
It seemed reasonable to presume that the direct oxidant of the SoxR cluster is O2− itself, since O2− is produced by redox-cycling agents and is a potent oxidant of some iron-sulfur clusters. Further, the induction of SOD is a prominent part of the response. Indeed, E. coli SOD mutants exhibited elevated expression of soxS and of members of the regulon (35). However, the degree of induction in these mutants was only about 20% of what is can be achieved with redox-cycling drugs, despite the fact that the concentration of O2− was high enough to inactivate several pathways that the SoxRS response has evolved to protect (36). Conversely, a plasmid that strongly overproduced SOD did not diminish the responsiveness of the SoxRS system to the redox-cycling drug paraquat (36, 37); thus toxic doses of O2− may be neither sufficient nor necessary to effectively induce the response. Further, recent reports indicate that redox-cycling drugs can activate SoxR in Pseudomonas aeruginosa under anaerobic conditions (38)--in which O2− formation is not possible--and that the genes that SoxR induces in Pseudomonads do not enhance O2− resistance (39). Thus the simple model that SoxR is a O2− sensor may be incomplete.
Because the redox state of the SoxR protein in vivo reflects the balance between oxidation and reduction, one possibility is that SoxR is continuously oxidized by molecular oxygen (and/or O2−), while redox-active antibiotics activate SoxR by interfering with its reduction. Roe and colleagues showed that the rsxABCDGE and rseC genes mediate SoxR reduction (40). They encode membrane-bound proteins that resemble members of the Rhodobacer capsulatus Rnf complex, which drives electrons onto nitrogenase. RsxC exhibits NADPH:cytochrome c oxidoreductase activity.
2.2. The OxyR and PerR regulators of responses to H2O2 stress
The oxyR gene was discovered by Christman et al. in a selection for Salmonella mutants that were hyperresistant to H2O2 (41). The OxyR protein belongs to the LysR family of transcription factors. Under activating conditions it binds as a tetramer near the −35 region of at least 20 regulon members (42) (Table 2), and it stimulates transcription through direct contact with RNA polymerase (43). Activation by H2O2 occurs through the oxidation of the Cys-199 residue to a sulfenic acid form. Although this point has been debated (44), most data indicate that a second cysteine residue (Cys-208) condenses with the sulfenic acid, forming a disulfide bond that locks the protein into a substantially altered conformation (45, 46). Because Cys-199 and Cys-208 are separated by 17 A in the crystal structure of the reduced enzyme (47), it is not obvious how these residues contact one another after oxidation. However, reduced Cys-199 sits in a hydrophobic pocket, and its oxidation to a larger, polar form may trigger its dissociation and a structural reorganization.
Oxidation of the purified protein by H2O2 is very rapid, with 100 nM creating a disulfide bond with a half-time of 30 s (48). It is appropriate that OxyR is calibrated to respond to such low doses, since sub-micromolar levels of intracellular H2O2 can cause substantial damage to DNA and enzymes (13, 14).
After H2O2 stress subsides, glutaredoxin 1 quickly reduces the Cys-199/Cys-208 disulfide bond through sulfur-exchange reactions (45). Since glutaredoxin 1 and glutathione reductase are themselves induced as part of the OxyR response, the system comprises a negative feedback cycle that allows induction to occur rapidly but be limited in magnitude.
While the OxyR system is widespread among bacteria, the gram-positive bacterium Bacillus subtilis uses a distinct H2O2 sensor (49). The PerR protein can bind a single ferrous iron atom, and in this form it is active as a transcriptional repressor. Upon exposure to H2O2, the iron atom is oxidized in a direct Fenton reaction that generates a ferryl and/or hydroxyl radical. This species covalently oxidizes one of the metal-coordinating histidyl residues (50), and this modification likely facilitates ferric iron dissociation and blocks the reloading of ferrous iron. The de-metallated protein lacks DNA-binding activity, allowing the induction of the genes whose transcription it normally blocks. Interestingly, many of the genes that PerR regulates are homologues or analogues of those in the OxyR regulon (Table 2).
3. Cellular defenses against oxidative stress
3.1. Scavenging systems
3.1.1. Scavengers of superoxide
The spontaneous dismutation of O2− is not sufficient to maintain low intracellular concentrations, because the reaction is second-order in O2− concentration and slows sharply as concentrations fall. Therefore, Gram-negative bacteria commonly synthesize both cytoplasmic and periplasmic isozymes of SOD as their front-line defense against O2−. E. coli contains two cytoplasmic SOD isozymes, one each of the manganese- and iron-cofactored types (MnSOD and FeSOD). It also secretes a single copper, zinc-cofactored enzyme (CuZnSOD, also called SodC) to the periplasm. Because O2− does not easily cross membranes at neutral pH (51, 52), O2− does not flow between these two compartments, and the physiological roles of the cytosolic and periplasmic enzymes can be considered separately. This arrangement is common, and it is ancestral to the mitochondrial MnSOD and cytoplasmic CuZnSOD of eukaryotes.
Although SOD exhibits a rate constant that approaches catalytic perfection, bacteria synthesize it in abundance. Exponentially growing E. coli contains about 50 μM total cytoplasmic SOD. Given that the rate of O2− formation is approximately 5 μM/s in these cells, the outcome is that the steady-state level of O2− is restricted to approximately 0.1 nM (53). This situation is unusual: the enzyme is four orders of magnitude more abundant than its substrate, and evolution has optimized the kinetics of an enzyme that must turn over only once every 10 s or so. Yet both abundance and catalytic proficiency are essential to keep O2− concentration low enough to protect vulnerable targets. Labile [4Fe-4S] enzymes are inactivated by O2− with a rate constant of 106–107 M−1 s−1, which means that even 10−10 M superoxide will inactivate them with a half-time of 30 min or so. Indeed, in experiments in which E. coli SOD titers were reduced by more than half, the activities of the [4Fe-4S] enzymes markedly declined, and growth defects emerged (36). The implication is that E. coli synthesizes just enough SOD activity to maintain the function of vulnerable enzymes in the face of endogenously generated O2−. In most microorganisms the synthesis of MnSOD is further elevated by an order of magnitude when the SoxRS system is activated by redox-cycling drugs.
Investigators are still working to identify the specific roles of periplasmic SOD. The housekeeping E. coli enzyme and the Salmonella SodCII evidently serve to defend unidentified periplasmic targets from O2− that leaks from respiratory-chain components on the outer aspect of the cytoplasmic membrane (6). However, periplasmic SOD evidently plays an additional role in several bacterial pathogens, whom it helps withstand the oxidative burst of host macrophages (54)
Initial surveys suggested that some obligately anaerobic bacteria lacked SOD (1)—raising the possibility that the lack of SOD was the feature that consigned them to anaerobic habitats. However, recent work has revealed that these organisms employ superoxide reductases, rather than dismutases, as scavengers (55, 56). The reason that obligate anaerobes cannot tolerate significant aeration is probably that molecular oxygen per se directly damages key enzymes; O2− scavengers cannot prevent that damage, but they can protect superoxide-sensitive enzymes whenever oxygenated waters briefly intrude into anaerobic habitats.
3.1.2. Scavengers of hydrogen peroxide
Hydrogen peroxide is scavenged in most organisms by peroxidases (eq. 3) and catalases (eq. 4):
| (3) |
| (4) |
The primary scavenger in E. coli and many bacteria is the peroxidiredoxin AhpCF, a two-component NADH peroxidase with a kcat/Km of 4 × 10e7 M−1 s−1 (57). H2O2 oxidizes Cys46 of AhpC to a sulfenic acid, which then condenses with Cys165 to form a Cys46-Cys165 disulfide bond. Exchange reactions with other AhpC cysteinyl residues re-reduce that bond at the expense of the creation of a second disulfide. This bond in turn is reduced upon binding of the NADH-reducible flavoprotein AhpF. In vivo the slow step in the overall process is likely to be the association and dissociation of the AhpF and AhpC proteins, rather than the initial reaction of AhpC with H2O2. That is fitting: under physiological conditions each protein need not turn over very frequently, but there is a premium on the rapidity with which each molecule of H2O2 is consumed by the first half of the reaction cycle. The activity of Ahp so high that although H2O2 is formed endogenously inside aerobic E. coli at a rate of about 15 μM/s, the steady-state concentration does not exceed 20 nM (58).
In vivo experiments show that when the extracellular concentration of H2O2 exceeds 20 μM, its influx into the cell causes the activity of Ahp to top out. Under these conditions the second half of the reaction cycle—reduction of oxidized AhpC by reduced AhpF—may be rate-limiting. Alternatively, the sulfenic acid of AhpC may be over-oxidized to a catalytically inactive sulfinic acid, as with peroxiredoxins found in mammalian systems (59). Inactivation in the face of high doses of H2O2 may be useful, as it keeps the enzyme from exhausting cellular NADH in a fruitless attempt to degrade overwhelming doses of H2O2.
When these higher doses of H2O2 saturate Ahp, the H2O2 concentration rises beyond the 0.1 μM threshold for OxyR activation (48, 58). Catalase is strongly induced, and it becomes the primary scavenging enzyme. The katG-encoded HPI of E. coli exhibits a high Km, so it is not saturated by even millimolar doses of H2O2 (60). Importantly, because catalase dismutates H2O2, its turnover rate is not restricted by the availability of reducing equivalents.
While H2O2 can passively diffuse across bacterial membranes at a substantial rate (58), that rate is easily matched by the rate of Ahp and/or catalase turnover, so that internal and external H2O2 concentrations do not equilibrate. When < 10 μM H2O2 is present in the bacterial environment, the intracellular concentration may be lower than the external concentration by an order of magnitude. The same is true of higher doses of H2O2, if the OxyR-regulated katG catalase is fully induced. Further, endogenously generated H2O2 is almost fully scavenged before it can cross the cytoplasmic membrane. Similar compartmentalization of H2O2 stress almost certainly pertains to eukaryotes, as reflected by the presence of peroxidases and/or catalases in each organelle.
3.2. Exclusion and export of redox-cycling antibiotics
Several members of the SoxRS system of E. coli collaborate to suppress the rate at which redox-cycling antibiotics accumulate inside the cell. When the MicF small RNA is expressed, it serves as an imperfect antisense RNA and occludes the ribosome-binding and translational start sites of the OmpF outer-membrane porin (61). By diminishing the abundance of this large porin, MicF slows the diffusion into the cell of these antibiotics (62).
SoxRS also induces the AcrAB drug-export system. The periplasmic (AcrA) and cytoplasmic membrane (AcrB) components interact with TolC to allow the pmf-driven efflux of a wide variety of antibiotics (63). The inclusion of the micF and acrAB genes within the SoxRS system strongly implies that this regulon evolved to defend the cell against oxidative stress that specifically arises from redox-cycling compounds.
3.3. Protecting iron-sulfur proteins
3.3.1. Repair processes
The oxidation of [4Fe-4S] dehydratase clusters, by either O2− or H2O2, leaves a [3Fe-4S]+ species; the polypeptide is undamaged. When O2− and/or H2O2 stress is terminated, intracellular enzyme activities rebound to their initial level even when new protein synthesis is blocked. The half-time of this process has been measured to be 3–5 minutes (36, 64). The mechanism of repair is unclear. In principle repair should require one-electron reduction of the cluster (to the [3Fe-4S]O state) and re-metallation by ferrous iron, and indeed the inactive dehydratases can be reactivated in vitro by incubation with dithiothreitol and ferrous iron. Neither the Isc nor the Suf systems, which are responsible for the de novo assembly of clusters in nascent polypeptides (65), are needed for cluster repair in vivo (66). New studies in E. coli and Salmonella have implicated the YtfE and YggX proteins in the process (67, 68), although the biochemical activities of both proteins remain unclear. Interestingly, YggX is induced as part of the SoxRS regulon (69).
3.3.2. Induction of oxidant-resistant isozymes
Fumarase A and aconitase B are among the dehydratases that are rapidly damaged by oxidants. It is therefore fitting that one component of the SoxRS response is the induction of fumarase C (70) and aconitase A (71), isozymes that are resistant to inactivation. Fumarase C is resistant because it does not use an iron-sulfur cluster in catalysis. In contrast, aconitase A has a cluster, and in fact the purified enzyme is acutely sensitive to oxidants. However, aconitase A retains activity in vivo and in extracts during exposure to a variety of oxidants, including H2O2 and O2− (72). The basis of protection is not clear; it might involve either a component that occludes the active site of the resting enzyme, or else aconitase A might be unusually amenable to the in vivo repair process. In any case, the synthesis of these enzymes comprises an elegant strategy to circumvent the effects of oxidative stress.
This strategy also poses a question: if an organism has oxidant-resistant enzymes in its repertoire, why not dispense with their vulnerable isozymes? The obvious hypothesis would be that the resistant enzymes are kinetically poorer catalysts, but this appears to be untrue (73, 74). Indeed, a fumarase C homologue is the exclusive isozyme in mammals. This conundrum remains unsolved. One idea that would explain the maintenance of aconitase B, however, is that its solvent exposure and general instability provide a mechanism through which the cell responds to iron starvation (75). A loss of activity may facilitate the accumulation and excretion of its substrate, citrate; citrate is an excellent iron chelator and serves as a siderophore with its own dedicated import system. Further, like the mammalian IRE-binding aconitase homologue, apo-aconitase is an RNA-binding protein that may control protein synthesis (76, 77).
3.2.3. Role of the Suf machinery
In E. coli the Isc system catalyzes the assembly of iron-sulfur clusters under routine growth conditions. Transcription is feedback-inhibited by the [2Fe-2S]-containing IscR repressor (78). The sufABCDSE operon encodes a second assembly system, and for a while its role was mysterious. The suf mutants do not exhibit growth or enzyme deficiencies during routine conditions. However, the operon is positively regulated by apo-IscR, implying that Suf provides a back-up system that compensates for conditional inadequacy of the primary Isc system (79, 80). Outten et al. found that the suf operon is repressed by Fur protein and induced by OxyR (81), which suggested that the Suf machine functions more efficiently than the Isc system when iron is scarce or H2O2 is present. Indeed, the suf mutants grow particularly poorly in iron-limited medium, and the activities of their Fe-S enzymes are lower than those of wild-type cells.
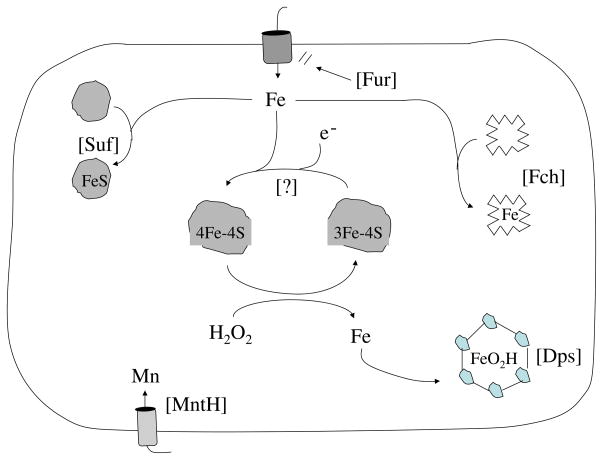
Further, suf mutants are hypersensitive to oxidizing conditions, failing to activate Fe-S enzymes during exposure to redox-cycling drugs (82) or exogenous H2O2 (S. Jang and J. A. Imlay, unpublished data). Evidently H2O2 disrupts function of the normal Isc machinery; the Suf machinery must rescue the cell because it is more resistant to oxidation (83). It seems plausible that H2O2 might disrupt nascent Fe-S clusters that are likely to be exposed on the IscU protein surface, much as it disrupts solvent-exposed FeS clusters within the active sites of dehydratases. Alternatively, the Suf machinery may be especially adept at functioning when intracellular free-iron concentrations are low, a situation that may arise when OxyR-induced Dps protein scavenges free iron to suppress Fenton chemistry (below). Ferrochetalase, the other enzyme that incorporates iron into cofactors, is also induced during H2O2 stress (Fig. 4).
Fig. 4.
Metal metabolism is altered during H2O2 stress. The Suf system restores cluster assembly, Dps sequesters unincorporated iron, Fur represses iron import, and ferrochetalase (Fch) ensures continued heme activation. The basis of protection by induced manganese import is uncertain.
3.4. DNA repair
DNA repair systems are essential for the aerobic lifestyle: mutants that lack both base-excision repair and recombinational repair functions--recA xthA and polA recB strains, for example--are viable only when they are cultured in anaerobic media (84, 85). Moreover, the mutation rates of wild-type E. coli are typically lower when cells are cultured anaerobically, and the spectrum of mutations shifts away from those that are associated with oxidative stress (86). These data indicate that in aerobic habitats oxidative damage is abundant enough that it is probably the predominant source of DNA damage and mutagenesis.
The details of the repair processes have been closely studied. Because DNA oxidation produces a wide variety of lesions, the base-excision pathways rely upon DNA glycosylases that scan the DNA for the absence of duplex integrity. E. coli endonuclease III and endonuclease VIII, for example, both recognize the disruption of the helix that results from saturation of pyrimidine residues. While single nth and nei mutants are not noticeably sensitive to H2O2, the double mutants are rapidly killed (87, 88). Endonucleases IV and III complete the repair process by incising the baseless sites, and they also address frank strand breaks that are produced by ribose oxidation. Thus nfo and xth mutants also are sensitive to peroxides (89, 90), and the double mutant is marginally viable in aerobic media. Interestingly, while eukaryotes do not contain genetic orthologues of these enzymes, they have functional analogues, and mutants that lack them are also hypersensitive to oxidants (91).
Guanine is a particular target of oxidation due to electron tunneling from this low-potential base to nearby base radicals (92). The most abundant product is 8-hydroxyguanine (8-oxoG). Its importance stems not only from the frequency of its formation but also from its ability to invert around its N-glycosidic bond and form a stable Hoogsteen mispair with adenine (93). The mispair is not easily detected by the proofreading system, and a high frequency of G → T transversions results. E. coli shares with many organisms several mechanisms to avoid this outcome. Formamidopyrimidine DNA glycosylase (Fpg, also known as MutM) is the principal glycosylase that excises oxidized purines prior to replication Tchou, 1991 #358]. If 8-oxoG is mis-replicated prior to excision, the 8-oxoG:A base pairs that are formed can be still corrected by MutY, an adenine DNA glycosylase that specifically operates upon these mispairs (94). Mutants lacking mutM and mutY are not appreciably sensitive to H2O2, but they exhibit high mutation rates in oxidizing environments. Finally, MutT is an enzyme that hydrolyzes 8-oxodGTP to 8-oxodGMP. It has a strong impact upon mutagenesis (95), indicating both that 8-oxodGTP can be formed by the direct oxidation of the deoxynucleotide pool and that DNA polymerase III will misincorporate this analogue.
Endonuclease IV is the only component of BER system that is induced in E. coli in response to oxidative stress (by SoxRS) (96); the remaining enzymes are constitutively expressed, presumably to cope with the DNA damage that is produced by endogenous H2O2. However, when DNA lesions persist, either because they are not suitable substrates for the BER system or because they are overwhelming in quantity, the SOS response is activated (97). This back-up system includes the induction of the UvrABC excinuclease that can remove glycosylase-resistant bulky lesions; inhibition of cell septation, to extend the window of opportunity for repair; stimulation of recombination activity to repair post-replication strand breaks; and the induction of alternative polymerases that can move the replication fork past persistent lesions. This regulon is induced in the period after the exposure of E. coli to substantial doses of H2O2, and a lexA3 mutation that blocks its induction results in hypersensitivity.
3.5. Protein repair
3.5.1. Mechanisms of polypeptide oxidation
The biological significance of protein oxidation, and of processes that may reverse it, is a point of uncertainty and great interest. Iron can bind avidly to polypeptides, and so Fenton chemistry is expected to target proteins as well as nucleic acids. In vitro model systems show that amino acid oxidations generate a wide variety of products (98); of these, carbonyls are the most easily quantified, and they are commonly used as proxies of protein oxidation inside cells. Consistent with expectation, protein carbonylation occurs most rapidly in vivo when bacteria are grown aerobically, when iron levels are high, or when cells are exposed either to H2O2 or to redox-cycling drugs (99). Some of the most profusely oxidized proteins include iron- or divalent cation-binding sites, including glutamate synthase, pyruvate kinase, and PtsI. However, other damaged proteins are ones that do not use prosthetic metals, raising the possibility that the injuries were catalyzed by iron atoms that bound adventitiously to the protein surface.
A key question is whether these reactions are broadly distributed over the total protein population, perhaps diminishing fitness due to a general accumulation of denatured and crosslinked proteins, or whether the reactions are sufficiently targeted to certain enzymes that specific pathways will fail. The answer rests upon two issues: the likelihood that iron occupies critical sites in a significant fraction of a given enzyme population, and the rate at which the bound iron atoms react with H2O2. In vitro protein oxidation systems have commonly used forcing circumstances to overcome these natural barriers. Efficient FeSOD inactivation, for example, requires supraphysiological levels of H2O2 (100), and isocitrate dehydrogenase is damaged only when enough iron is added to out-compete manganese for the metal binding site (101). To date no metabolic pathway failures in moderately stressed cells have been linked to these types of metal-catalyzed oxidation reactions.
The oxidation of most amino acids appears to be irreversible, and proteolytic degradation is the likely recourse for the cell. In contrast, methionine sulfoxide reductases and disulfide reducing systems are universal among organisms. Indeed, Cys and Met are likely to be disproportionate targets of Fenton chemistry in proteins, both because they exhibit very high rate constants for reaction with hydroxyl radicals, and also because they may tunnel electrons to nearby amino-acid radicals (102). However, the more common suspicion is that H2O2 directly oxidizes these residues without resort to Fenton chemistry.
At present, however, the evidence is mixed. The rate constants for the oxidation of typical cysteine and methionine residues by H2O2 are quite low (2–20 M−1 s−1 and 0.01 M−1 s−1) (103, 104), indicating that they are unlikely to react with physiological doses of H2O2. In contrast, OxyR, the organic hydroperoxide sensor OhrR, and peroxiredoxins achieve rate constants in the 107 M−1 s−1 range (48, 57). In these proteins the pKa of the reactive cysteine is lowered through its interaction with a cationic partner, and it seems likely that a nearby Thr/Tyr/His residue (105–107) polarizes the peroxyl bond and ultimately protonates the hydroxyl leaving group.
Therefore, one might anticipate that adventitious oxidation reactions would specifically damage enzymes that have similar active-site arrangements. Glyceraldehyde-3-phosphate dehydrogenase, for example, has received attention because it is inactivated when eukaryotic cells are stressed by moderate doses (ca. 50 μM) of exogenous H2O2 (108). The enzyme employs a cysteine thiolate as a nucleophile; a histidine residue opposes the substate binding site, where in the course of normal catalysis it stabilizes and then protonates the reaction intermediate (109). However, despite these general similarities to the architecture of H2O2-responsive proteins, the enzyme reacts with H2O2 with a rate constant that is still only ~ 60 M−1 s−1 (110). Similarly low values appear to pertain to other proteins that are either activated or inactivated during acute H2O2 stress—including eukaryotic tyrosine phosphatases (111), bacterial methionine synthases (112), and the Hsp33 protein chaperone of E. coli (113). Thus these proteins are not significantly oxidized by the sub-micromolar doses of H2O2 that trigger the OxyR response and damage Fe-S enzymes.
3.5.2. Methionine sulfoxide reductase
Most organisms contain two methionine sulfoxide reductases, each of which specifically reduce one of the two enantiomers of the oxidized residue (114):
| (5) |
Despite the poor reactivity of methionine with H2O2, the disruption of msrA conferred vulnerability to growth inhibition by H2O2 and/or paraquat in E. coli (115), Erwinia (116) and yeast (117). Further, mutants exhibited defects in protein secretion that were tracked to the oxidation of a critical methionine in the signal recognition particle complex (118). In these latter experiments exogenous oxidants were not provided, indicating that endogenous, unidentified oxidants continually damage proteins. The msr genes are not induced in E. coli by either OxyR or by SoxRS. Perhaps repair systems must continue to function after the inducing oxidant has dissipated, and so it would be non-optimal for their synthesis to be controlled in the same manner as that of scavenging enzymes.
3.5.3. The reduction of disulfide bonds
Thioredoxins and glutaredoxins are the primary reductants of intracellular disulfide bonds (119). These small proteins were originally identified by their roles in electron delivery to ribonucleotide reductase and PAPS reductase, two enzymes that employ sulfur chemistry to reduce unusual substrates. Several other thiol-based reductases were subsequently discovered in E. coli—methionine sulfoxide reductase, arsenate reductase, and DsbD of the periplasmic disulfide reducing system.
Most workers presumed that thioredoxins and glutaredoxins also serve to reduce adventitious protein disulfide bonds that are formed during oxidative stress. This idea is strongly supported by the discovery that the E. coli OxyR regulon activates synthesis of glutathione reductase, glutaredoxin 1, and thioredoxin 2 during exposure to H2O2. Similarly, the Yap-1 system of S. cerevisiae triggers induction of glutathione synthesis, glutathione reductase, glutaredoxin 2, thioredoxin reductase, and thioredoxin 2 (120).
H2O2 unquestionably triggers protein disulfide formation in eukaryotic cells. However, the oxidation may not be direct, since the same products might be generated when protein thiols undergo exchange reactions with the oxidized glutathione that accumulates as a product of glutathione peroxidase. In bacteria and yeast, where Gpx is a minor scavenger or absent altogether, the evidence of protein thiol oxidation by physiological doses of H2O2 is not strong. High H2O2 doses have typically been employed in proteomics studies that detect oxidized proteins (121, 122). One wonders, then, whether glutaredoxins and thioredoxins are actually needed only when H2O2 stress is of such long duration as to compensate for the sluggish reactivity of protein thiols; whether they repair a small cohort of extremely reactive thiolate enzymes that have so far escaped detection; whether thiolate oxidations are primarily driven by reactions with oxygen or hydroxyl radicals, rather than H2O2; or whether disulfide stress arises in natural environments from a different types of stressor, mimicked by thiol agents such as diamide. In fact, many bacteria—including Bacillus, Mycobacteria, Rhodobacter, and Streptomyces—do not include glutaredoxins or thioredoxins in their H2O2-inducible regulons (123–125). Instead, for example, Streptomyces controls synthesis of its thioredoxins with an anti-sigma factor protein that is activated when sulfur exchange reactions create a disulfide bond (126); notably, this system is easily triggered by diamide but requires millimolar doses of H2O2. Thus at the moment the importance of thiol oxidation in H2O2 toxicity, and the role that redoxins play in H2O2 defenses, remain puzzling questions.
3.6. Controls on the levels of unincorporated iron
Since hydroxyl radical is formed by reaction between H2O2 and ferrous iron, its toxicity is limited when their concentrations are minimized. The primary control of iron homeostasis in most bacteria is mediated by Fur protein, a transcription factor that is activated by the binding of ferrous iron (127). Metallated Fur binds to the promoter regions of operons that encode iron-import proteins, thereby effecting feedback control.
EPR measurements indicated that the level of unincorporated iron in lab-cultured E. coli ranges from about 20 micromolar in defined medium to 100 micromolar in iron-rich complex medium (13, 25). This free-iron pool presumably represents iron that is in transit as well as iron that binds adventitiously to the surfaces of biomolecules. When the fur gene was mutated, these iron levels rose 5- to 10-fold. The fur mutants exhibited high rates of mutagenesis (128) and were proportionately more sensitive to DNA damage by exogenous H2O2 (25). Further, fur recA mutants, which are defective at repairing oxidative DNA damage, were found to be inviable in aerobic media (128). These results certify the important role that Fur protein plays in minimizing DNA oxidation.
Interestingly, expression of the fur gene is induced by both the OxyR and SoxS proteins (129). Positive control by OxyR is evidently required because H2O2 tends to inactivate the ferrous-Fur complex (72), perhaps by oxidation of the ferrous iron cofactor. When the OxyR binding site upstream of fur was disrupted, cells that were exposed to micromolar H2O2 inappropriately induced the Fur regulon, over-imported iron, and suffered debilitating amounts of DNA damage.
Interestingly, the toxicity that E. coli fur mutants experience from excessive iron import can be compensated for by the engineered over-synthesis of the classic iron-storage protein, ferritin (128). Ferritin is strongly induced by OxyR and PerR systems in some H2O2-stressed bacteria (130, 131); Dps, an alternative iron-storage protein (132, 133), is activated in many others (134). Dps appears to use H2O2 rather than molecular oxygen as the electron acceptor during iron oxidation, perhaps because this arrangement offers a mechanism whereby the storage function may be deactivated when H2O2 stress has ended. E. coli dps mutants are hypersensitive to H2O2 (135); in fact, E. coli catatalase/peroxidase mutants are unable to grow in aerobic environments if the dps gene is inactivated, as endogenous H2O2 creates overwhelming amounts of DNA damage (13).
3.7. How does manganese protect microbes against oxidative stress?
In early surveys of the distribution of SOD among microorganisms, Lactobacillus plantarum stood out as an oxygen-tolerant organism that lacked superoxide dismutase activity (136). However, L. plantarum did accumulate enormous concentrations of intracellular manganese—ca. 20 mM—when grown in manganese-supplemented medium. When manganese was not provided, growth suffered. The implication was that manganese might chemically scavenge O2−, making the synthesis of an enzyme unnecessary.
Consistent with this proposal, manganese(II) reduces O2− with a rate constant in the range of 106 M−1 s−1 (137), which in principle means that millimolar concentrations would have the clearance activity of micromolar SOD. In fact, manganese supplements can diminish or eradicate the phenotypic deficits of SOD mutants of E. coli, B. subtilis, and S. cerevisiae (138–140).
However, the regulation of manganese importers is slightly at odds with this model: the proton-driven manganese importer MntH is induced (by OxyR) when E. coli is exposed to H2O2 (141), not O2−—and the ATP-driven importer MntABC is induced in Stapholococcus aureus under the same circumstances by its PerR system (142). Further, mutations that eliminate these systems sensitize a variety of microbes to H2O2 (143) (141). In explaining these data, some workers have speculated that Mn may scavenge H2O2 as well as O2−, since carbonate-bound manganese shows a catalase-like activity in vitro (144).
However, recent work has clouded this picture, too. Several of the bacteria that accumulate high intracellular concentrations of manganese also contain substantial titers of SOD and catalase (145). And it is inherently discomforting to posit that cells employ a non-enzymatic system to catalyze a chemical reaction—an arrangement that would be rare, if not unprecedented.
Recently we found that mntH mutations prevent the aerobic growth of E. coli peroxidase/catalase mutants (A. Anjem and J.A. Imlay, unpublished data): in these cells the endogenous H2O2 is toxic unless manganese can be imported. Direct measurements show that manganese import does not affect the rate at which H2O2 effluxes from these cells; thus the role of manganese during oxidative stress is likely to be something other than the scavenging of O2− or H2O2. One possibility is that the high (millimolar) intracellular concentrations of manganese allow it to outcompete unincorporated iron for binding to adventitious cation-binding sites on proteins and/or nucleic acids. Manganese and iron tolerate similar coordination environments, and they are known to compete for binding in vivo even to fairly selective sites in metalloenzymes such as superoxide dismutases (146), isocitrate dehydrogenase (101), Fur protein, and PerR protein (147). By quantitatively outcompeting iron for non-specific divalent sites, manganese could prevent site-specific Fenton reactions that might otherwise damage these biomolecules (148). This model has not yet been tested.
Prospects
Twenty years ago our understanding of intracellular oxidative stress was in its infancy. The existence of superoxide dismutases and catalases implied that ROS are formed endogenously and have toxic effects, and it was just becoming clear these enzymes were regulated as parts of larger defensive responses. Since then our understanding has substantially broadened, and details have come into focus. The view from here suggests that we are about at the half-way point: We know of mechanisms that form reactive oxygen species, but not the specific reactions that are primarily responsible in vivo; we have pinpointed several oxidative injuries that cause growth disruptions, but several phenotypes remain unexplained; and we have deduced the purposes of about half of the oxidant-inducible genes, but many of the rest are ORFs of unknown function. Currently genetic and physiological approaches are illuminating new truths—and debunking old assumptions—at an accelerating rate. The burgeoning ability to study oxidative stress in a wide variety of microbial and non-microbial organisms will almost certainly challenge our instinct to generalize the data from E. coli, but this work will also underscore those aspects of oxidative stress that are truly universal.
Supplementary Material
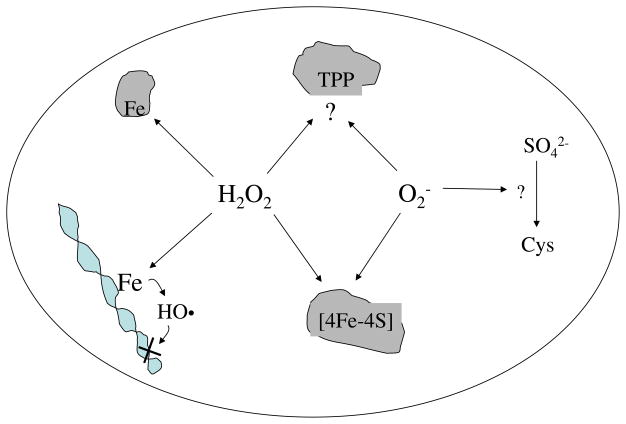
Fig. 3.
Known mechanisms by which H2O2 and O2− injure cells include Fenton-mediated damage to proteins and DNA, the oxidation of solvent-exposed [4Fe-4S] clusters, apparent inhibition of transketolase, and disruption of the sulfur assimilatory pathway. The details of inactivation of transketolase and of sulfur metabolism remain unclear.
Acknowledgments
I apologize to the many investigators whose specific results could not be cited due to space limitations but whose work framed the questions and ideas that are discussed in this review. J.A.I. is supported by NIH GM49640.
Key terms and definitions
- Reactive oxygen species (ROS)
Used generally to refer to activated derivatives of molecular oxygen, including singlet oxygen, superoxide, hydrogen peroxide, hydroxyl radical, hypohalous acids, and peroxynitrite
- Fenton reaction
Electron transfer from ferrous iron to hydrogen peroxide, generating a hydroxyl radical as an oxidizing product
- Unincorporated (“free”) iron
Functionally, iron within a cell that has not been incorporated into hemes, iron-sulfur clusters, or high-affinity mononuclear sites within proteins. Therefore, this term includes iron in transit and iron that is loosely associated with biomolecules
- Redox-cycling drugs
Toxic chemical agents, including viologens, soluble quinones, and phenazines, that penetrate bacterial cells and catalyze electron transfer from redox enzymes to molecular oxygen, generating superoxide and hydrogen peroxide as products
- Peroxiredoxins
A widely distributed family of peroxidases that divalently reduce hydrogen peroxide and/or organic hydroperoxides through an active site thiolate, generating a sulfenic acid intermediate
Abbreviations and acronyms
- SOD
superoxide dismutase
- Ahp
alkylhydroperoxide reductase
Literature cited
- 1.McCord JM, Keele BB, Jr, Fridovich I. Proc Natl Acad Sci USA. 1971;68:1024–7. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ligeza A, Tikhonov AN, Hyde JS, Subczynski WK. Biochim Biophys Acta. 1998;1365:453–63. doi: 10.1016/s0005-2728(98)00098-x. [DOI] [PubMed] [Google Scholar]
- 3.Massey V, Strickland S, Mayhew SG, Howell LG, Engel PC, et al. Biochem Biophys Res Commun. 1969;36:891–7. doi: 10.1016/0006-291x(69)90287-3. [DOI] [PubMed] [Google Scholar]
- 4.Messner KR, Imlay JA. J Biol Chem. 1999;274:10119–28. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 5.Seaver LC, Imlay JA. J Biol Chem. 2004;279:48742–50. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- 6.Korshunov S, Imlay JA. J Bacteriol. 2006;188:6326–34. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Proc Natl Acad Sci USA. 1990;87:6181–5. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsaneva IR, Weiss B. J, Bacteriol. 1990;172:4197–205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielski BHJ, Richter HW. J Amer Chem Soc. 1977;99:3019. [Google Scholar]
- 10.Fee JA. Trends Biochem Sci. 1982;7:84–6. [Google Scholar]
- 11.Sawyer DT, Valentine JS. Acc Chem Res. 1981;14:393–400. [Google Scholar]
- 12.Carlioz A, Touati D. EMBO J. 1986;5:623–30. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, You X, Imlay JA. Proc Natl Acad Sci USA. 2005;102:9317–22. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang S, Imlay JA. J Biol Chem. 2007;282:929–37. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo CF, Mashino T, Fridovich I. J Biol Chem. 1987;262:4724–7. [PubMed] [Google Scholar]
- 16.Flint DH, Tuminello JF, Emptage MH. J Biol Chem. 1993;268:22369–76. [PubMed] [Google Scholar]
- 17.Farr SB, D’Ari R, Touati D. Proc Natl Acad Sci USA. 1986;83:8268–72. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin DE, Hollstein M, Christman MF, Schwiers EA, Ames BN. Proc Natl Acad Sci USA. 1982;79:7445–9. doi: 10.1073/pnas.79.23.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henle ES, Han Z, Tang N, Rai P, Luo Y, et al. J Biol Chem. 1999;274:962–71. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- 20.Imlay JA, Chin SM, Linn S. Science. 1988;240:640–2. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 21.Walling C. Acc Chem Res. 1975;8:125–31. [Google Scholar]
- 22.Rush JD, Maskos Z, Koppenol WH. FEBS Letts. 1990;261:121–3. [Google Scholar]
- 23.Park S, Imlay JA. J Bacteriol. 2003;185:1942–50. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liochev SI, Fridovich I. Free Rad Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 25.Keyer K, Imlay JA. Proc Natl Acad Sci USA. 1996;93:13635–40. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benov L, Fridovich I. J Biol Chem. 1999;274:4202–6. doi: 10.1074/jbc.274.7.4202. [DOI] [PubMed] [Google Scholar]
- 27.Chang EC, Kosman DJ. J Biol Chem. 1989;264:12172–8. [PubMed] [Google Scholar]
- 28.Hassan HM, Fridovich I. J Biol Chem. 1977;252:7667–72. [PubMed] [Google Scholar]
- 29.Pomposiello PJ, Bennik MH, Demple B. J Bacteriol. 2001;183:3890–902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding H, Demple B. Proc Natl Acad Sci USA. 1997;94:8445–9. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaudu P, Moon N, Weiss B. J Biol Chem. 1997;272:5082–6. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo E, Ding H, Demple B. Cell. 1997;88:121–9. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 33.Eiamphungporn W, Charoenlap N, Vattanaviboon P, Mongkolsuk S. J Bacteriol. 2006;188:8669–73. doi: 10.1128/JB.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi K, Tagawa S. J Biochem (Tokyo) 2004;136:607–15. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- 35.Liochev SI, Benov L, Touati D, Fridovich I. J Biol Chem. 1999;274:9479–81. doi: 10.1074/jbc.274.14.9479. [DOI] [PubMed] [Google Scholar]
- 36.Gort AS, Imlay JA. J Bacteriol. 1998;180:1402–10. doi: 10.1128/jb.180.6.1402-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liochev SI, Fridovich I. Arch Biochem Biophys. 1992;294:138–43. doi: 10.1016/0003-9861(92)90147-o. [DOI] [PubMed] [Google Scholar]
- 38.Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, Newman DK. Mol Microbiol. 2006;61:1308–21. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 39.Park W, Pena-Llopis S, Lee Y, Demple B. Biochem Biophys Res Commun. 2006;341:51–6. doi: 10.1016/j.bbrc.2005.12.142. [DOI] [PubMed] [Google Scholar]
- 40.Koo MS, Lee JH, Ray SY, Yeo WS, Lee JW, et al. EMBO J. 2003;22:2614–22. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christman MF, Morgan RW, Jacobson FS, Ames BN. Cell. 1985;41:753–62. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 42.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, et al. J Bacteriol. 2001;183:4562–70. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao K, Fujita N, Ishihama A. Mol Microbiol. 1993;7:859–64. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 44.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, et al. Cell. 2002;109:383–96. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 45.Zheng M, Aslund F, Storz G. Science. 1998;279:1718–21. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 46.Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, et al. Nat Struct Mol Biol. 2004;11:1179–85. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 47.Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, et al. Cell. 2001;105:103–13. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 48.Aslund F, Zheng M, Beckwith J, Storz G. Proc Natl Acad Sci USA. 1999;96:6161–5. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bsat N, Chen L, Helmann JD. J Bacteriol. 1996;178:6579–86. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JW, Helmann JD. Nature. 2006;440:363–7. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 51.Lynch RE, Fridovich I. J Biol Chem. 1978;253:4697–9. [PubMed] [Google Scholar]
- 52.Korshunov SS, Imlay JA. Mol Microbiol. 2002;43:95–106. doi: 10.1046/j.1365-2958.2002.02719.x. [DOI] [PubMed] [Google Scholar]
- 53.Imlay JA, Fridovich I. J Biol Chem. 1991;266:6957–65. [PubMed] [Google Scholar]
- 54.De Groote MA, Granger D, Xu Y, Campbell G, Prince R, et al. Proc Natl Acad Sci USA. 1997;94:13997–4001. [Google Scholar]
- 55.Jenney FE, Jr, Verhagen MFJM, Cui X, Adams MWW. Science. 1999;286:306–9. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 56.Lombard M, Fontecave M, Touati D, Niviere V. J Biol Chem. 2000;275:115–21. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- 57.Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, et al. Biochemistry. 2005;44:10583–92. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seaver LC, Imlay JA. J Bacteriol. 2001;183:7182–9. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poole LB. Arch Biochem Biophys. 2005;433:240–54. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 60.Hillar A, Peters B, Pauls R, Loboda A, Zhang H, et al. Biochemistry. 2000;59:5868–75. doi: 10.1021/bi0000059. [DOI] [PubMed] [Google Scholar]
- 61.Delihas N, Forst S. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- 62.Zhang A, Wassarman KM, Rosenow C, Tiaden BC, Storz G, et al. Mol Microbiol. 2003;50:1111–24. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 63.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Curr Opin Struct Biol. 2004;14:741–7. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Gardner PR, Fridovich I. J Biol Chem. 1992;267:8757–63. [PubMed] [Google Scholar]
- 65.Barras F, Loiseau L, Py B. Adv Microb Physiol. 2005;50:41–101. doi: 10.1016/S0065-2911(05)50002-X. [DOI] [PubMed] [Google Scholar]
- 66.Djaman O, Outten FW, Imlay JA. J Biol Chem. 2004;279:44590–9. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 67.Justino MC, Almeida CC, Teixeira M, Saraiva LM. J Biol Chem. 2007;282:10352–9. doi: 10.1074/jbc.M610656200. [DOI] [PubMed] [Google Scholar]
- 68.Gralnick J, Downs D. Proc Natl Acad Sci USA. 2001;98:8030–5. doi: 10.1073/pnas.151243198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pomposiello PJ, Koutsolioutsou A, Carrasco D, Demple B. J Bacteriol. 2003;185:6624–32. doi: 10.1128/JB.185.22.6624-6632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liochev SI, Fridovich I. Proc Natl Acad Sci USA. 1992;89:5892–6. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cunningham L, Gruer MJ, Guest JR. Microbiology. 1997;143:3795–805. doi: 10.1099/00221287-143-12-3795. [DOI] [PubMed] [Google Scholar]
- 72.Varghese S, Wu A, Park S, Imlay KRC, Imlay JA. Mol Microbiol. 2007;64:822–30. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flint DH, Allen RM. Chem Rev. 1996;96:2315–34. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- 74.Jordan PA, Tang Y, Bradbury AJ, Thomson AJ, Guest JR. Biochem J. 1999;344:739–46. [PMC free article] [PubMed] [Google Scholar]
- 75.Varghese SM, Tang Y, Imlay JA. J Bacteriol. 2003;185:221–30. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang Y, Guest JR. Microbiology. 1999;145:3069–79. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 77.Alen C, Sonenshein AL. Proc Natl Acad Sci USA. 1999;96:10412–7. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, et al. Proc Natl Acad Sci USA. 2001;98:14895–900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yeo WS, Lee JH, Lee KC, Roe JH. Mol Microbiol. 2006;61:206–18. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 80.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. Mol Microbiol. 2006;60:1058–75. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 81.Outten FW, Djaman O, Storz G. Mol Microbiol. 2004;52:861–72. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 82.Nachin L, Loiseau L, Expert D, Barras F. EMBO J. 2003;22:427–37. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. J Biochem (Tokyo) 2004;136:199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 84.Morimyo M. J Bacteriol. 1982;152:208–14. doi: 10.1128/jb.152.1.208-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imlay JA, Linn S. J Bacteriol. 1986;166:519–27. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakai A, Nakanishi M, Yoshiyama K, Maki H. Genes to Cells. 2006;11:767–78. doi: 10.1111/j.1365-2443.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- 87.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. J Bacteriol. 1997;179:3773–82. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, et al. J Bacteriol. 1997;179:3783–5. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Demple B, Halbrook J, Linn S. J Bacteriol. 1983;153:1079–82. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. J Bacteriol. 1986;168:1120–7. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Demple B, DeMott MS. Oncogene. 2002;21:8926–34. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- 92.Candeias LP, Steenken S. J Amer Chem Soc. 1993;115:2437–40. [Google Scholar]
- 93.Hogg M, Wallace SS, Doublie S. Curr Opin Struct Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 94.Michaels ML, Cruz C, Grollman AP, Miller JH. Proc Natl Acad Sci USA. 1992;89:7022–5. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fowler RG, Schaaper RM. FEMS Microbiol Rev. 1997;21:43–54. doi: 10.1111/j.1574-6976.1997.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 96.Chan E, Weiss B. Proc Natl Acad Sci USA. 1987;84:3189–93. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Imlay JA, Linn S. J Bacteriol. 1987;169:2967–76. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davies MJ. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 99.Dukan S, Nystrom T. J Biol Chem. 1999;274:26027–32. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- 100.Beyer WF, Jr, Fridovich I. Biochemistry. 1987;26:1251–7. doi: 10.1021/bi00379a008. [DOI] [PubMed] [Google Scholar]
- 101.Murakami K, Tsubouchi R, Ogawa MF, Yoshino M. Arch Microbiol. 2006;186:385–92. doi: 10.1007/s00203-006-0153-1. [DOI] [PubMed] [Google Scholar]
- 102.Dean RT, Fu S, Stocker R, Davies MJ. Biochem J. 1997;324:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Winterbourn CC, Metodiewa D. Free Rad Biol Med. 1999;27:322–8. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 104.Griffiths SW, Cooney CL. Biochemistry. 2002;41:6245–52. doi: 10.1021/bi025599p. [DOI] [PubMed] [Google Scholar]
- 105.Hofmann B, Hecht H-J, Flohe L. Biol Chem. 2002;383:347–64. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 106.Kona J, Brinck T. Org Biomol Chem. 2006;4:3468–78. doi: 10.1039/b604602a. [DOI] [PubMed] [Google Scholar]
- 107.Soonsanga S, Fuangthong M, Helmann JD. J Bacteriol. 2007 doi: 10.1128/JB.00879-07. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grant CM, Quinn KA, Dawes IW. Mol Cell Biol. 1999;19:2650–6. doi: 10.1128/mcb.19.4.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Talfournier F, Colloc’h N, Mornon JP, Branlant G. Eur J Biochem. 1998;252:447–57. doi: 10.1046/j.1432-1327.1998.2520447.x. [DOI] [PubMed] [Google Scholar]
- 110.Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, et al. FEBS J. 2007;274:212–26. doi: 10.1111/j.1742-4658.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 111.Denu JM, Tanner KG. Biochemistry. 1998;37:5633–42. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 112.Hondorp ER, Matthews RG. PLoS Biol. 2004;2:1738–1753. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jakob U, Muse W, Eser M, Bardwell JC. Cell. 1999;96:341–52. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 114.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, et al. J Biol Chem. 2001;276:48915–20. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 115.Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, et al. J Bacteriol. 1995;177:502–7. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hassouni ME, Chambost JP, Expert D, Gijsegem FV, Barras F. Proc Natl Acad Sci USA. 1999;96:887–92. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, et al. Proc Natl Acad Sci USA. 1998;95:14071–5. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ezraty B, Aussel L, Barras F. Biochim Biophys Acta. 2005;1703:221–9. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 119.Ritz D, Beckwith J. Annu Rev Biochem. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 120.Carmel-Harel O, Storz G. Annu Rev Microbiol. 2000;54:439–61. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 121.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. J Biol Chem. 2006;281:10420–30. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 122.Leichert LI, Jakob U. PLoS Biol. 2004;2:1723–37. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li K, Hein S, Zou W, Klug G. J Bacteriol. 2004;186:6800–8. doi: 10.1128/JB.186.20.6800-6808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Mol Microbiol. 2001;42:1007–20. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 125.Helmann JD, Wu MFW, Gaballa A, Kobel PA, Morshedi MM, et al. J Bacteriol. 2003;185:243–53. doi: 10.1128/JB.185.1.243-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kang J-G, Paget MSB, Seok Y-J, Hahn M-Y, Bae J-B, et al. EMBO J. 1999;18:4292–8. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Neilands JB. Arch Biochem Biophys. 1993;302:1–3. doi: 10.1006/abbi.1993.1172. [DOI] [PubMed] [Google Scholar]
- 128.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. J Bacteriol. 1995;177:2305–14. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng M, Doan B, Schneider TD, Storz G. J Bacteriol. 1999;181:4639–43. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. Infect Immun. 2001;69:3744–54. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rocha ER, Smith CJ. Microbiology. 2004;150:2125–34. doi: 10.1099/mic.0.26948-0. [DOI] [PubMed] [Google Scholar]
- 132.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, et al. J Biol Chem. 2002;277:27689–96. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 133.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 134.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. Mol Microbiol. 1994;13:265–72. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 135.Martinez A, Kolter R. J Bacteriol. 1997;179:5188–94. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Archibald FS, Fridovich I. J Bacteriol. 1981;146:928–36. doi: 10.1128/jb.146.3.928-936.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gray B, Carmichael AJ. Biochem J. 1992;281:795–802. doi: 10.1042/bj2810795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Al-Maghrebi M, Fridovich I, Benov L. Arch Biochem Biophys. 2002;402:104–9. doi: 10.1016/S0003-9861(02)00065-6. [DOI] [PubMed] [Google Scholar]
- 139.Inaoka T, Matsumura Y, Tsuchido T. J Bacteriol. 1999;181:1939–43. doi: 10.1128/jb.181.6.1939-1943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sanchez RJ, Srinivasan C, Munroe WH, Wallace MA, Martins J, et al. J Biol Inorg Chem. 2005;10:912–23. doi: 10.1007/s00775-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 141.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. Mol Microbiol. 2000;36:1085–100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 142.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, et al. Mol Microbiol. 2002;44:1269–86. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 143.Que Q, Helmann JD. Mol Microbiol. 2000;35:1454–88. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 144.Berlett BS, Chock PB, Yim MB, Stadtman ER. Proc Natl Acad Sci USA. 1990;87:389–93. doi: 10.1073/pnas.87.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, et al. FEMS Micro Rev. 2005;29:361–75. doi: 10.1016/j.femsre.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 146.Beyer WF, Jr, Fridovich I. J Biol Chem. 1991;266:303–8. [PubMed] [Google Scholar]
- 147.Herbig AF, Helmann JD. Mol Microbiol. 2001;41:849–59. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 148.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, et al. PLoS Biol. 2007;5:769–79. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.