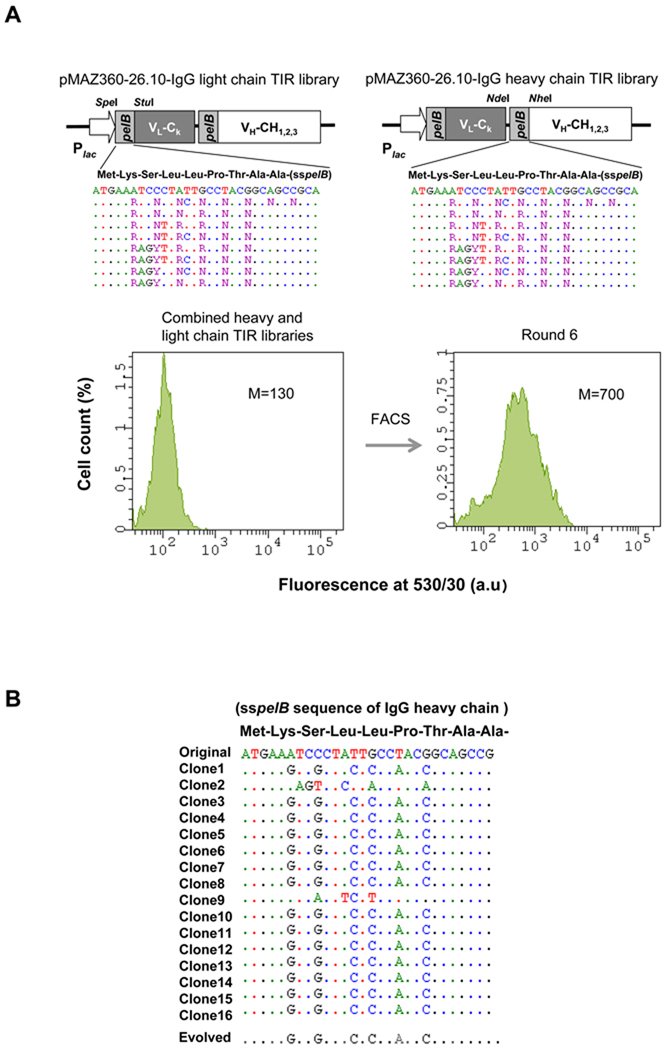
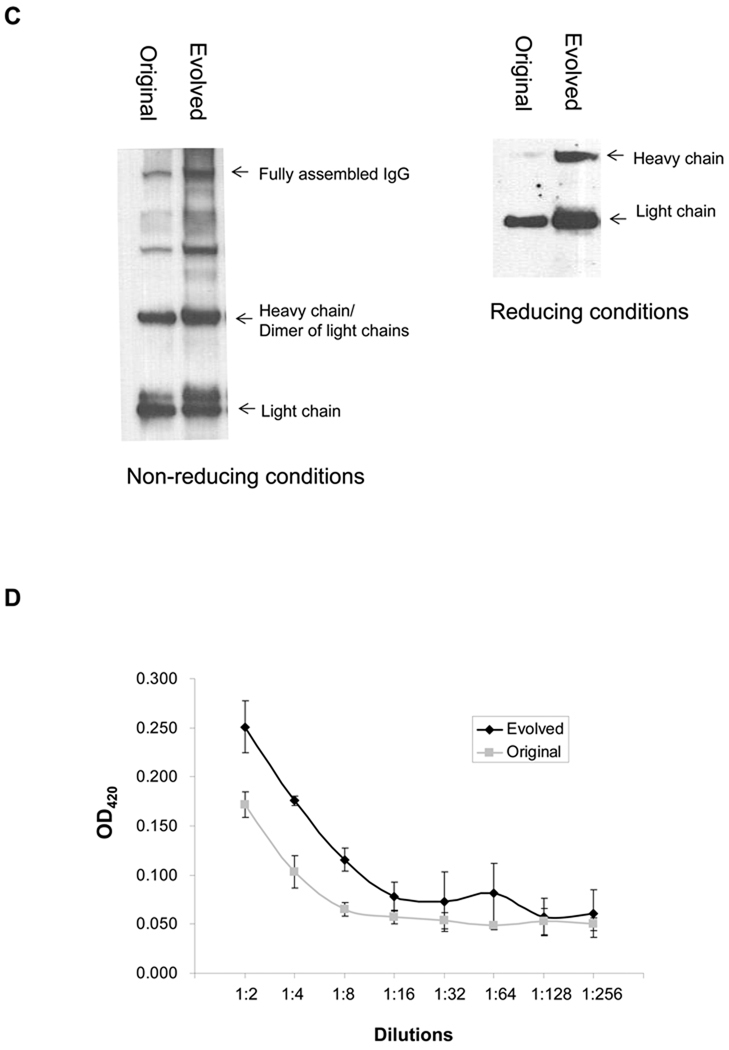
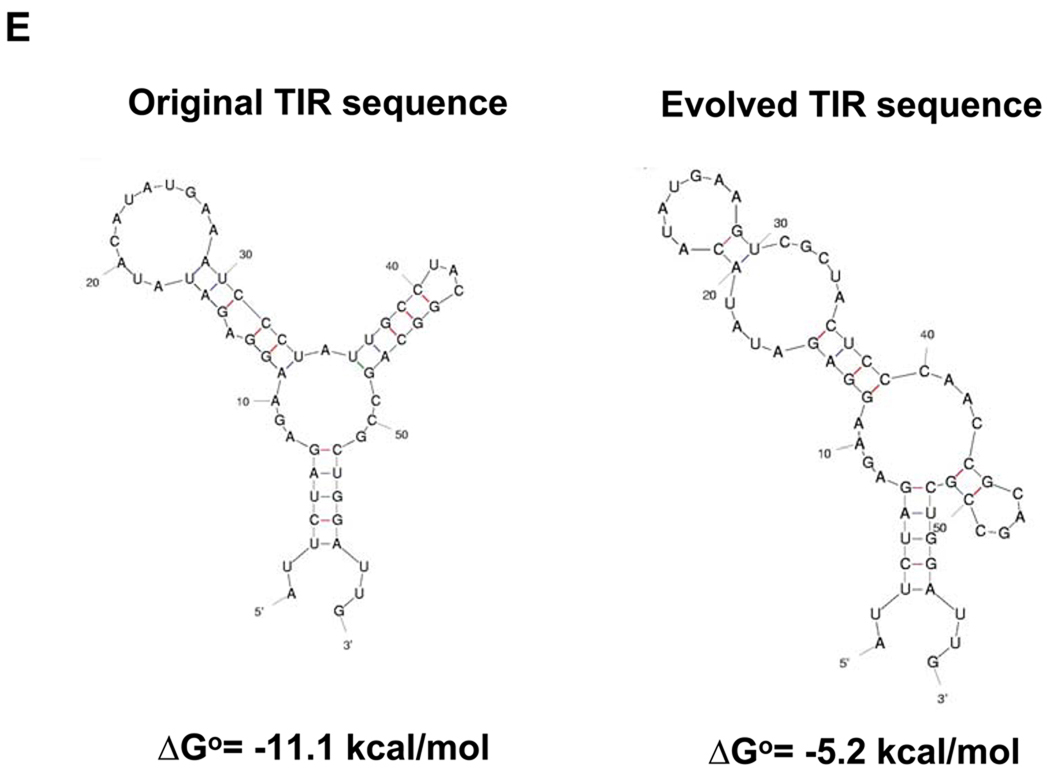
Figure 3. Screening of a Library for Optimized Translational Initiation Region (TIR) Sequences.
(A) Construction of TIR libraries for light and heavy chain genes and screening by FACS. M: mean fluorescence intensity, sspelB: pelB signaling sequence (B) Nucleotide sequences of the heavy chain TIRs of the isolated clones. (C) Western blotting of JUDE-1 cells carrying the pMAZ360-26.10-IgG vector either with the original or the selected heavy chain TIR sequences under non-reducing (left) and reducing (right) conditions. All lanes were loaded with total protein corresponding to the same number of cells as judged by OD600. (D) ELISA determination of antigen-binding activity in cell lysates from mutant clones. Experiments were carried out in triplicate and the error bars correspond to one standard deviation from the mean values. (E) Prediction of mRNA secondary structure for the original and the evolved heavy chain TIR sequences using the Mfold software (Zuker, 2003). ΔGo: Gibbs free energy of unfolding.