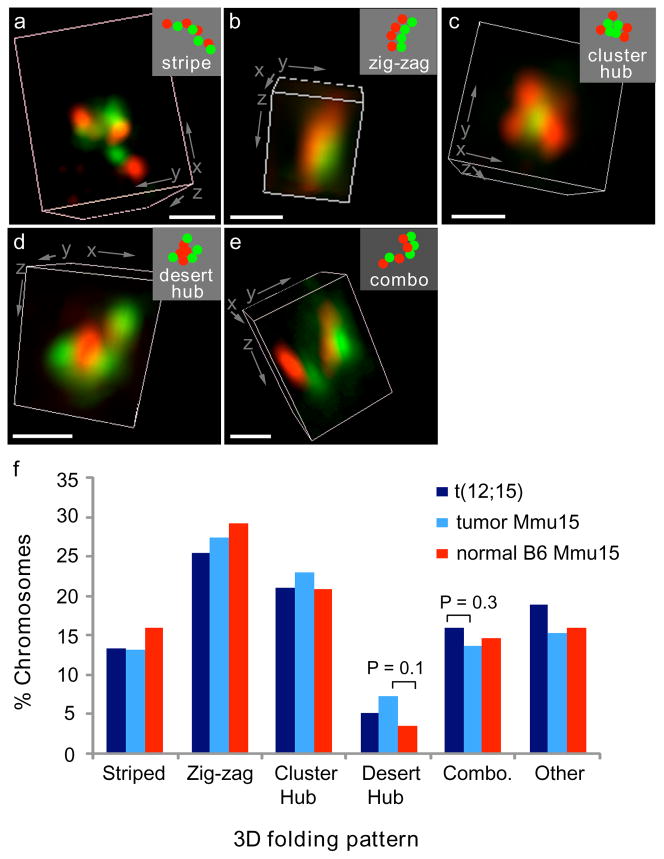
Fig. 4. Patterns of higher-order chromatin folding are not significantly altered by translocation or tumorigenesis.
(a–e) The 7 Mb of Mmu15 distal to Myc and/or the t(12;15) breakpoint were labeled by FISH as in Fig. 3. The relative positions of its gene clusters (green) and deserts (red) were classified as one of five morphological patterns: (a) striped fiber, (b) zigzags, (c) gene cluster hub, (d) gene desert hub, or (e) a combination (combo) of two of the other four patterns. 3D renderings of deconvolved images are shown. Insets (top right) are schematic diagrams of probe patterns. Bars, 500 nm. (f) Scoring the frequency of 3D folding patterns of normal Mmu15 (N = 234) and t(12;15) (N = 232) from the three tumors analyzed indicated no significant difference between the translocated and normal chromosomes (P ≥ 0.3, χ2 tests). Normal pro-B cells from B6 mice (N = 144) also had a similar distribution of folding patterns (P ≥ 0.1, χ2 tests).