Figure 2. Schematic of a typical GWAS design.
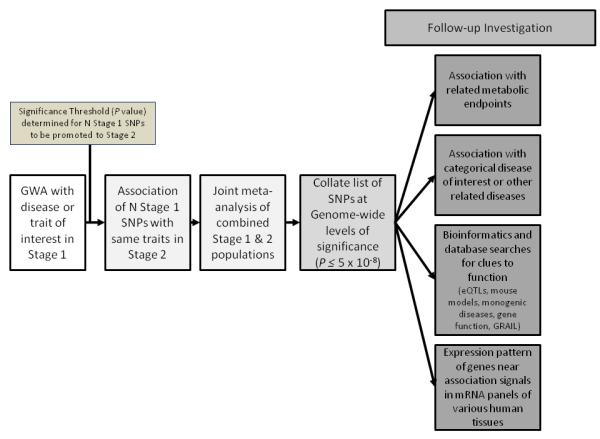
The general design of GWAS starts with a stage 1 (discovery) cohort. The top SNPs are promoted to the stage 2 (replication) cohort based on 1) a significance threshold that is usually dictated by pragmatic considerations, 2) whether there is prior knowledge of association between the disease the variant, and 3) whether an association is biologically plausible. The successfully replicated SNPs are meta-analyzed in the combined stage 1 and stage 2 cohorts. The SNPs that reach levels of genome-wide significance (P=5×10−8) are explored further using the functional analysis techniques listed.
