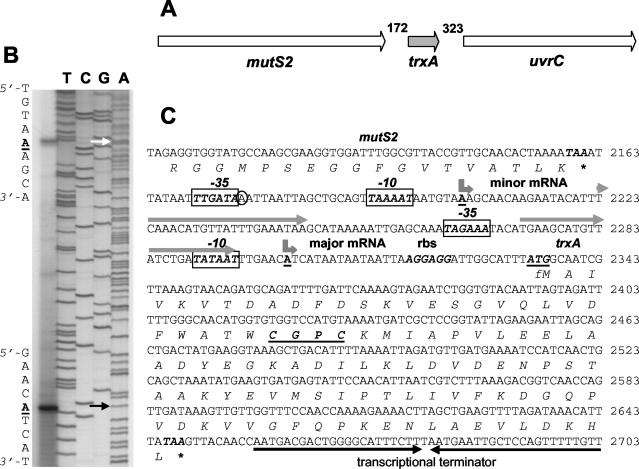
FIG. 1.
Chromosomal organization, primer extension analysis, and sequence of the S. aureus Oxford trxA gene. (A) The gene organization is identical to that found in all of the S. aureus strains referred to in this work; the numbering of nucleotides in intergenic regions is for S. aureus Mu50, accession no.. NC_002758. The ≈5.3-kbp DNA region containing trxA and flanking genes is shown. Gene designations: mutS2 encodes a mismatch repair ATPase; trxA encodes thioredoxin; uvrC encodes a subunit of the endonuclease nucleotide excision repair system. (B) Total RNA was isolated from aerobic cultures of S. aureus Oxford. Primer extension was carried out as described in Materials and Methods, and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primers. Arrows point to the A nucleotides of the two trxA transcription start sites. (C) Nucleotide sequence of the trxA promoter (numbering according to GenBank AJ223480). Transcription start points are shown by bent arrows above the underlined boldface A nucleotides. The trxA ATG translational start codon (underlined) and its ribosome-binding site are shown in boldface italic letters. Putative −10 and −35 hexamer sequences are shown as boxed boldface italic letters. Two direct repeats are shown by grey arrows above the nucleotide sequences. The TAA stop codons of the mutS2 and trxA genes are marked by an asterisk. A putative rho-independent transcriptional terminator is indicated by two solid inverted arrows below the nucleotide sequence.