FIGURE 2.
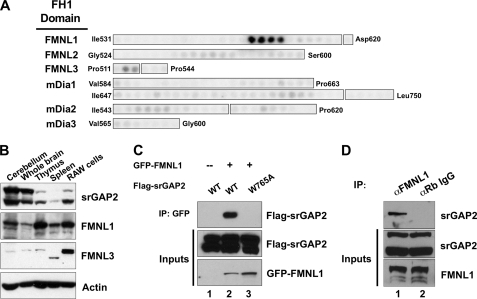
srGAP2 forms a complex with FMNL1. A, shown is a peptide array analysis of the binding specificity of the srGAP2 SH3 domain for the FH1 domains of several formins. The beginning and ending amino acid positions indicate regions synthesized as 20-mer peptides offset every 3 amino acids. Binding was detected by overlay of γ-32P-radiolabeled GST-srGAP2 SH3. Results suggested the srGAP2 SH3 domain prefers peptides within the FMNL1 FH1 domain. B, expression distribution of srGAP2, FMNL1, and FMNL3 were determined by Western blot analysis using specific antibodies. Actin was used as a control for loading. Each panel is labeled to the right, and each tissue or cell line is labeled above each panel. RAW cells are a macrophage-derived cell line. C, full-lengths rGAP2 and FMNL1 interact by co-immunoprecipitation (IP). Cells were transfected with GFP-FMNL1 alone (lane 1) or with FLAG-srGAP2 (lane 2) or FLAG-srGAP2 with a point mutation in the SH3 domain (tryptophan 765 to alanine; lane 3). Immunoprecipitation of FMNL1 co-precipitated wild type srGAP2 (lane 2, top panel), but GFP alone or the SH3 mutant of srGAP2 did not (lanes 1 and 2, top panel). Immunoblot analysis of extracts indicated equal levels of srGAP2 in all three lysates (middle panel) and similar levels of FMNL1 (bottom panel). D, endogenous FMNL1 and srGAP2 form a complex in cells. Immunoprecipitation of FMNL1 co-precipitates srGAP2 from HeLa cells (lane 1), whereas α-rabbit IgG (negative control) does not (lane 2).