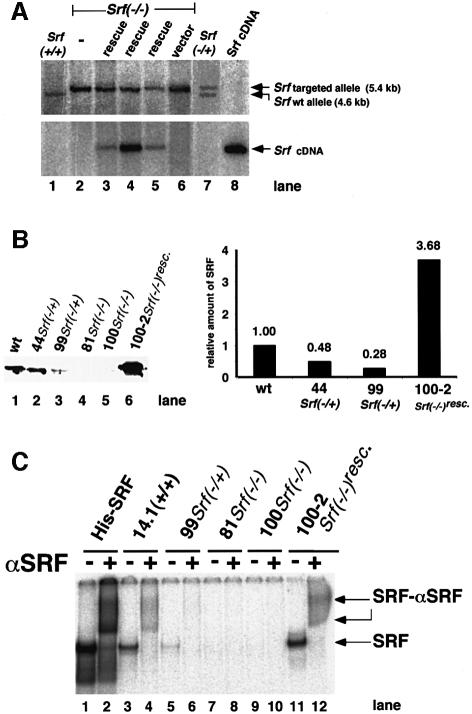
Fig. 1. Genotypes and SRF protein activities of ES cells mutated at the Srf locus. (A) Genotyping by Southern blotting of ES cell lines used in this study. Genomic DNAs from the ES cell lines E14.1 Srf+/+ (lane 1), 226 Srf–/+ (lane 7) and 226-100 Srf–/– (lane 2), 226-100 Srf–/– plus human Srf cDNA expression construct pSGSRF1 (rescue) (lanes 3–5) or 226-100 Srf–/– plus an empty vector (vector) (lane 6) were digested with BglII, electrophoresed and blotted. Filters were successively hybridized with either a murine Srf probe discriminating between wild-type and targeted alleles (top panel) or a human Srf probe detecting the integrated human Srf cDNA (bottom panel). The plasmid pSGSRF1 was used as a hybridization control (Srf cDNA) (lane 8). Positions of the BglII fragments representing the wild-type allele (4.6 kb) or the targeted allele (5.4 kb) are indicated. Lanes 3, 4 and 5 represent the ‘rescue’ lines 226-100-2, 226-100-37 and 226-100-42, respectively. (B) Western analysis of SRF protein in extracts of ES cell lines used in this study. The blot (left) and quantification of the density of the bands on it (right) are shown. No specific SRF western signal was observed in the homozygous Srf–/– ES cell lines. (C) SRF-mediated SRE binding activities in extracts of ES cell lines used in this study. A radiolabeled c-fos SRE oligonucleotide probe was incubated with recombinant His-tagged SRF (lanes 1 and 2) or protein extracts from ES cells of the different genotypes, as indicated. Even-numbered lanes represent extracts with added polyclonal anti-SRF serum. SRF-containing DNA–protein complexes were supershifted by this antiserum. No SRF-containing complexes were detected in extracts from Srf–/– ES cells (lanes 7–10). Lanes 11 and 12 contained 4 µg of protein extract only, whereas 20 µg of protein was used for other cell extracts.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.