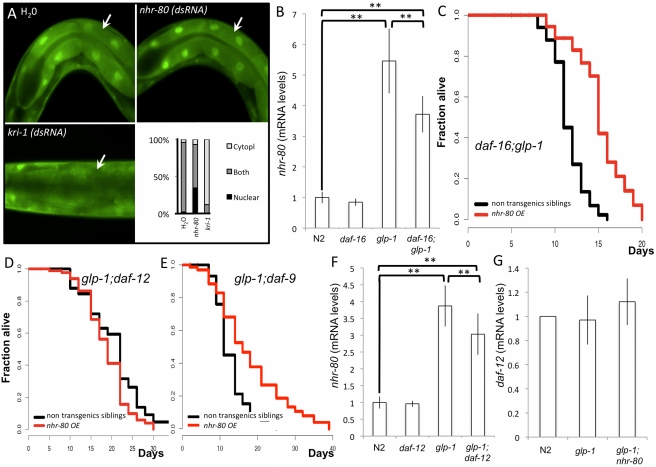
Figure 5. NHR-80 can function in the absence of daf-16 but requires daf-12.
(A) DAF-16 localization is not affected by nhr-80. Using the CF1935 strain (daf-16(mu86)I;glp-1(e2141ts)III; muIs109), we assessed the localization of DAF-16::GFP by fluorescent microscopy. Fertile daf-16(mu86);glp-1(e2141ts);muIs109 [Pdaf-16::gfp::daf-16] mutant animals were injected with water (control), kri-1, or nhr-80 dsRNA (1 mg/ml) as previously described [4]. Progeny of injected animals were assayed for DAF-16 localization in intestinal cells, indicated by a white arrow, using a fluorescent microscope (Axioplan, Zeiss). DAF-16::GFP is strongly nuclear in the progeny of water injected (upper left panel) or nhr-80 dsRNA injected animals (upper right panel), while it remains cytoplasmic when kri-1 dsRNA was injected (lower left panel). To ensure that injection of nhr-80 ds RNA was efficient, lifespan of the progeny was monitored and was shortened as expected (unpublished data). Images are magnified 630-fold. The lower right panel shows a quantification of these results. Percent of animals showing either nuclear, cytoplasmic localization, or both is depicted on the graph (50, 29, and 25 animals were analyzed for water control, nhr-80 (dsRNA), and kri-1 (dsRNA), respectively). (B) nhr-80 mRNA levels are increased in glp-1(e2141ts) and in daf-16(mu86);glp-1(e2141ts) mutants as measured by qRT-PCR (5.6- and 3.7-fold increase; Wilcoxon rank-sum test p value <0.05 for both strains when compared with N2). When daf-16(mu86);glp-1(e2141ts) mutants are compared to glp-1(e2141ts) mutants, there is a 1.5-fold decrease (Wilcoxon rank-sum test p value <0.05). Error bars are standard deviation (*p<0.1, **p<0.05, ***p<0.01, Wilcoxon rank-sum test). This suggests that nhr-80 mRNA levels respond to glp-1(e2141ts) mutants and that part of this response is daf-16 independent. A control experiment shows that nhr-80 is not under the control of daf-16 in the wild type background (Wilcoxon rank-sum test p value is non-significant). (C) nhr-80 overexpression (lynEx[(nhr-80p::nhr-80::gfp);myo-2p::DsRed]) increases the lifespan of daf-16(mu86);glp-1(e2141ts) mutant animals (p<0.0001, 38% extension in mean lifespan) but (D) fails to increase the lifespan of glp-1(e2141ts);daf-12(rh61rh411) double mutants (MLS of 18 and 19.5 d for glp-1(e2141ts);daf-12(rh61rh411) mutants carrying the transgene or not; p = 0.17). (E) Finally, nhr-80 overexpression increases the lifespan of glp-1(e2141ts);daf-9(rh50) double mutants (MLS of 15 and 10 d for glp-1(e2141ts);daf-9(rh50) mutants carrying the transgene or not, respectively; p<0.0001, 50% extension in mean lifespan). Thus, while NHR-80 can partially bypass the need for DAF-16, its longevity function requires the presence of DAF-12, but not of DAF-9. Lifespan analyses were performed independently at least twice. The p values were calculated using the log rank (Mantel-Cox) analyses. (F) nhr-80 mRNA levels are up-regulated in glp-1(e2141ts) and in glp-1(e2141ts);daf-12(rh61rh411) mutant animals (3.87- and 3.01-fold induction, respectively, compared to N2 treated similarly; Wilcoxon rank-sum test p value <0.05 for both; error bars are standard deviation). When compared to glp-1(e2141ts) mutants, nhr-80 mRNA levels are decreased 1.2-fold in glp-1(e2141ts);daf-12(rh61rh411) (Wilcoxon rank-sum test p value <0.05; error bars are standard deviation). nhr-80 induction does not strictly require DAF-12 but is modulated by DAF-12. (G) daf-12 mRNA levels are unaffected in glp-1(e2141ts) and in glp-1(e2141ts);nhr-80(tm1011) mutant animals (1.07- and 1.10-fold induction, respectively, compared to N2 treated similarly; the Wilcoxon rank-sum test p value is non-significant; error bars are standard deviation).