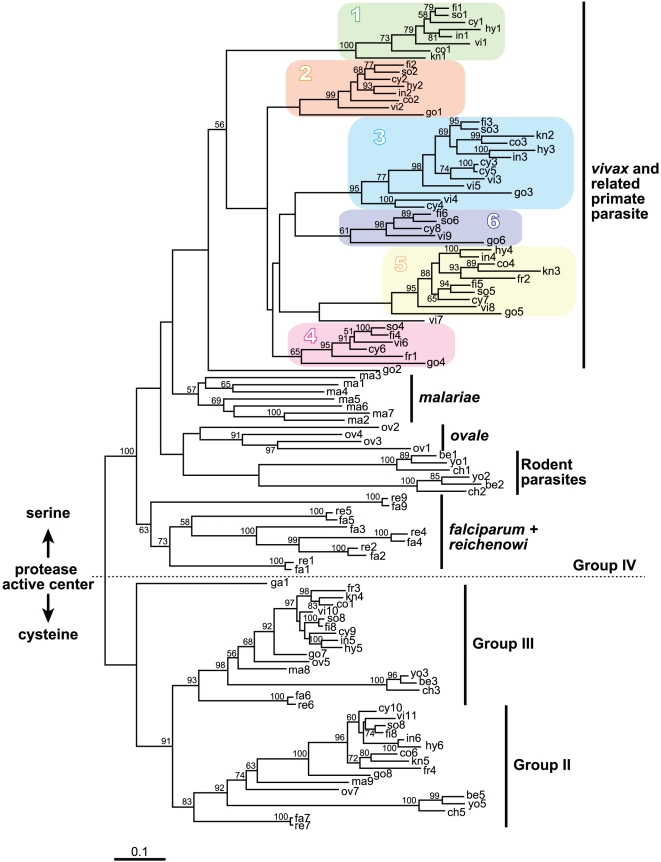
Figure 2. The maximum likelihood (ML) phylogenetic tree of Plasmodium SERA genes.
This unrooted tree was constructed from 115 SERA genes (encompassing Groups II to IV, see Figure 1) using 570 amino acid positions under the JTT + Γ (eight categories) model (α = 1.15) with 500 heuristic replicates. Bootstrap proportions >50% are shown along nodes. Groups II - III and Group IV are cysteine-type and serine-type SERA genes, respectively. Note that Pga-SERA1 (ga1) is an offshoot of Groups II and III SERA genes, suggesting the occurrence of a common ancestor, leading to Pga-SERA1 (ga1) and a common ancestor of Group II and Group III. In P. vivax and P. vivax-related monkey malaria parasite species, the six clades are color-boxed. Pgo-SERA1 (go1) and Pgo-SERA5 (go5) were grouped, despite low bootstrap values, into Clade 2 and Clade 5 respectively, because these genes showed common features to each clade in exon/intron structure and/or gene array.