Figure 2.
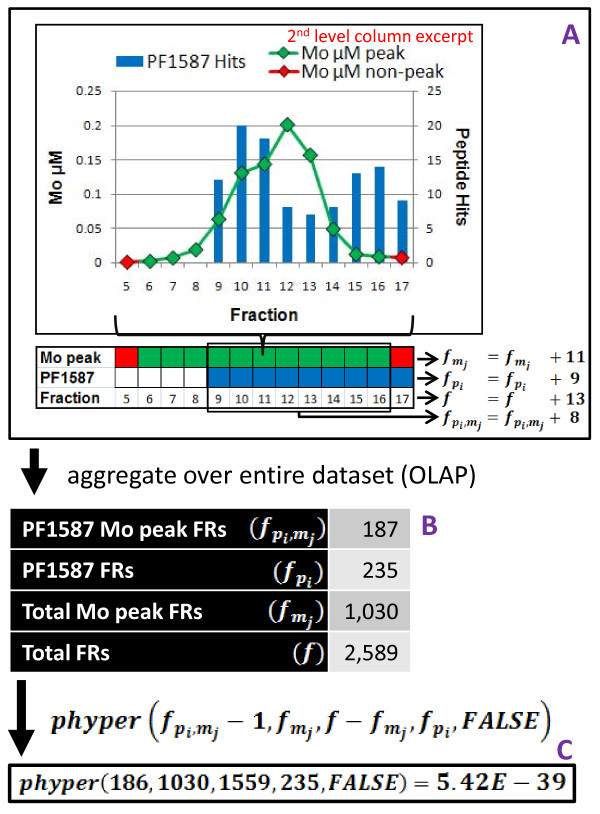
GMPA score (Global Metal Protein Association) Calculation. A, B and C illustrate the calculation with data for pi = PF1587 and mj = Mo, arrows represent generic steps in the calculation. A) Peptide counts for each protein (per fraction) are reduced to Boolean values (present/not present-shown as blue/white cells respectively). Whether a fraction is part of a metal peak or not is already a Boolean value (green/red cells). B) The "present/not present" values are counted across all fractions in the data set. C) The GMPA score is calculated from these values using the hypergeometric distribution and is roughly a p-value: how likely is a given protein to have been seen in metal peak fractions as many times as it was (or more) assuming an equal likelihood for the protein to have been observed in any fraction and given the number of metal peak fractions, protein fractions, and total fractions.
