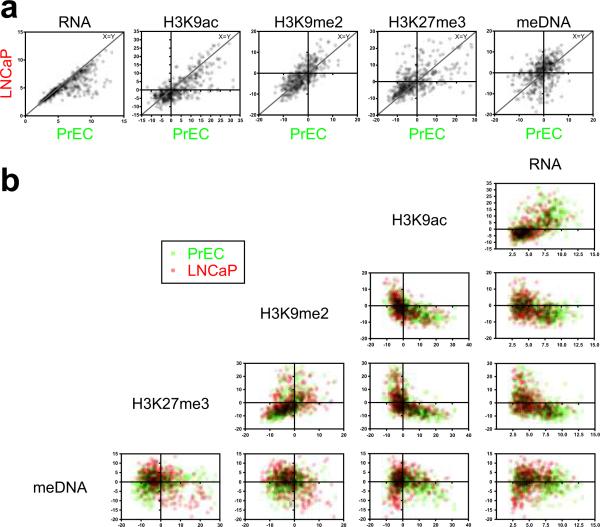
Fig. 5. Scatter plots of epigenetic marks in all LRES genes.
RNA signals, as well as summarised ChIP and MeDIP signals were compared for all LRES genes. (For each gene, the sum was determined of the MAT scores at −2kb, −1kb, TSS and +1kb relative to its transcription start site. Transparent data points are shown and overlaying signals have been multiplied to facilitate a comprehensive interpretation.) (a) Scatter plots comparing RNA, H3K9ac, H3K9me2, H3K27me3 and DNA methylation (meDNA) signals in PrEC and LNCaP cells. Data points close to the line x = y reflect genes that have not changed their mark between the cell lines. A Wilcoxon signed rank test indicated significant depletions in RNA and H3K9ac signals in LNCaP compared to PrEC cells, while H3K27me3 and meDNA levels were overall increased (all P values were <0.0005). (b) Matrix scatter plots of signals within each cell line (PrEC cells: green; LNCaP cells: red). H3K9ac signals are high when RNA levels are high. H3K9me2 and H3K27me3 signal are only high when H3K9ac or RNA levels are low or off. H3K9me2 and H3K27me3 reveal a positive correlation, while especially in LNCaP genes H3K27me3 and DNA methylation signal show a negative correlation. Horizontal and vertical lines in each plot indicate y = 0 and x = 0, respectively.