Fig. 2.
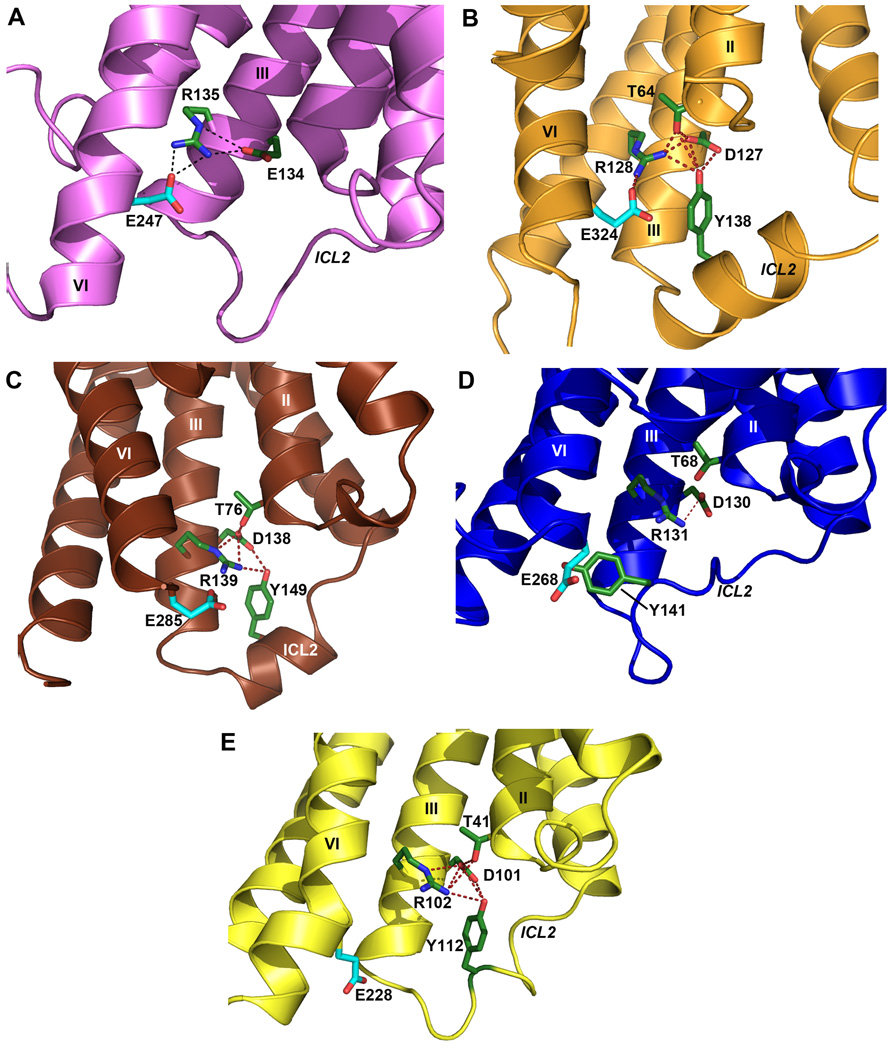
Conformation of ICL2 and ionic lock motif in D3R and other GPCR structures. As also seen in (A) the inactive Rhodopsin structure (PDB ID: 1U19), the conserved ionic lock motif D[E]RY is in a “locked” conformation in (B) the D3R structure, i.e. with a salt bridge formed between Arg1283.50 and Glu3246.30. In addition, the side chain of Tyr138 in the ICL2 α-helix of the D3R is inserted into the seven-TM bundle forming hydrogen bonds with Thr642.39, Arg1283.50 and Asp1273.49 (distances 3.0 Å, 3.2 Å and 3.2 Å, respectively), potentially stabilizing the ionic lock. There is no salt bridge between Arg3.50-Glu6.30 (and hence the “ionic lock” is “broken”) in other crystal structures of GPCR shown in panels (C) β1AR (PDB ID: 2VT4), (D) β2AR (PDB ID: 2RH1), (E) A2AAR (PDB ID: 3EML). In both the β1AR and A2AAR structures, however, the corresponding Tyr residue in ICL2 that aligns to Tyr138 in D3R forms two hydrogen bonds with the Asp3.49 and Arg3.50 side chains even in the absence of the closed ionic lock conformation. Salt bridges are shown as red dashed lines, and hydrogen bond interactions are shown as blue dashed lines.