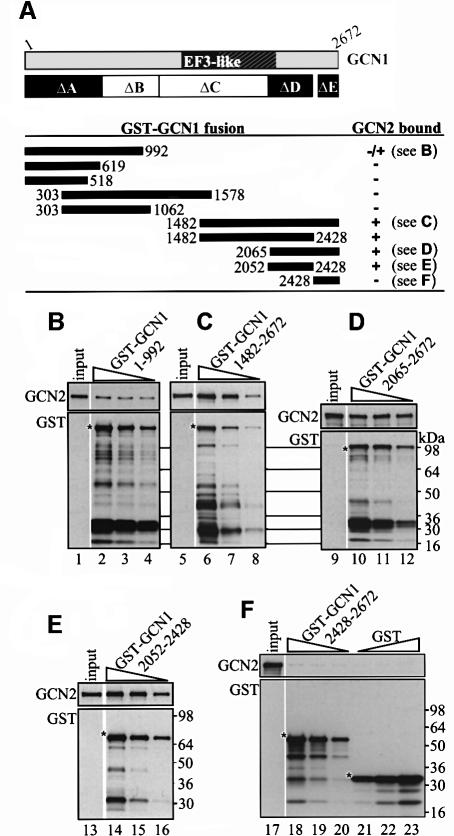
Fig. 4. A C-terminal segment of GCN1 (region D) is sufficient for GCN2 binding in vitro. GCN1 fragments fused to GST were expressed in E.coli, immobilized on glutathione–Sepharose beads and incubated with 1 mg whole-cell extract from yeast gcn1Δ strain H2556 containing the high copy GCN2 plasmid pAH15. After washing, the beads were subjected to immunoblot analysis using GCN2 and GST antibodies to measure the amounts of bound proteins. (A) A schematic of GCN1 is shown, labeled as in Figure 1C, and the regions highlighted in black are necessary for GCN2 binding in vivo (see Figure 3). Below are shown schematically the GCN1 fragments contained in the GST–GCN1 fusions (solid rectangles with numbers indicating amino acid positions) and a qualitative summary of their GCN2 binding activities based on the results in (B–F) and data not shown. The plasmids encoding the GST–GCN1 fusions were (from top to bottom): pES131-2, pES145-1, pES141-2, pES99-3, pES84-2, pES79-1, pES86-1, pES123-B1 and pES136-1. (B–F) Results for each of the indicated GST–GCN1 fusions of three binding assays with different concentrations of fusion protein (each one differing by a factor of 2 from the other) and a sample containing 10% of the yeast extract (input) are shown. The upper and lower panels show the results of immunoblot analysis obtained with GCN2 and GST antibodies, respectively. An asterisk indicates the predicted full-length GST fusion proteins.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.