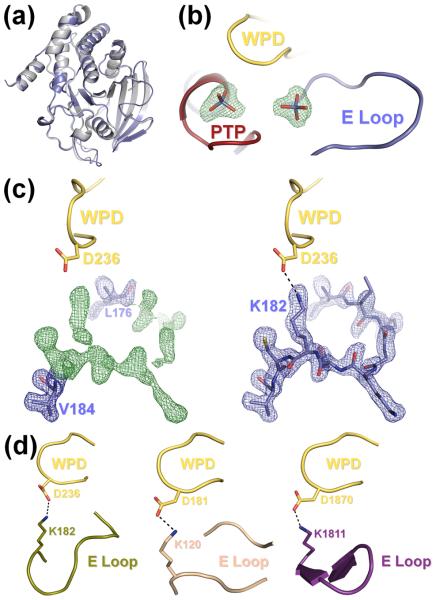
Figure 2.
(a) The S72D mutation does not alter the overall structure of HePTP. Superposition of the structures of HePTP0 (blue) and WT HePTP (grey; PDB ID: 1ZC010). The structures are nearly identical (RMSD < 0.5 Å over all mainchain atoms). (b) HePTP0 contains sulfate bound at the active site and at a secondary binding site. mFO – DFC electron density map (green mesh; contoured at 3.0 σ to 1.90 Å) of the HePTP active site and interface of the PTP, WPD and E loops. Both sites contained well-defined, tetrahedral electron density (≥9.9 σ) from the early stages of refinement. (c) Visualization of the E loop in HePTP0. Left panel, 2mFO – DFC electron density map (blue mesh; contoured at 1.0 σ to 1.90 Å) and mFO – DFC electron density map (green mesh; contoured at 3.0 σ to 1.90 Å) of residues Leu176 and Val184 refined at an occupancy of 1.0 (blue, stick representation) with E loop residues 177–183 omitted from refinement. Right panel, 2mFO – DFC electron density map (blue mesh; contoured at 1.0 σ to 1.90 Å) of E loop residues 176–184 refined at an occupancy of 1.0 (blue). E loop residue Lys182 forms a hydrogen bond (illustrated by a dashed line) with the catalytic Asp236 of the WPD loop (yellow). Protein models prepared using PyMOL.36 (d) The WPD-E loop interaction is conserved across multiple, distinct PTP structures. Although the conformation of the E loop is different between HePTP0 (blue, right panel of (c)), HePTP C270S* (olive, left panel), PTP1B (beige, middle panel; PDB ID: 2B4S), and RPTPβ (purple, right panel: PDB ID: 2I4E), the catalytic aspartate of the WPD loop (yellow) in each of these structures forms a similar hydrogen bond with the conserved lysine of the E loop (illustrated by a dashed line).