Figure 3.
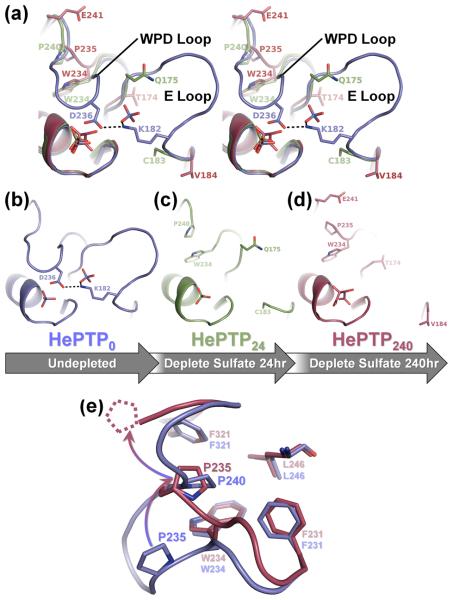
Depletion soaking induces coordinated movement of the HePTP WPD and E loops. (a) Stereo image of the superposition of the PTP, WPD and E loops of HePTP0 (blue), HePTP24 (green) and HePTP240 (red). In HePTP0 (blue), the WPD and E loops are well-ordered, and WPD loop residue Asp236 hydrogen bonds with E loop residue Lys182 (illustrated as a dashed line). In crystals depleted of sulfate for 24 hours (i.e. HePTP24; green) and 240 hours (i.e. HePTP240; red), the WPD and E loops simultaneously become disordered. Residues in HePTP24 and HePTP240 at ordered-to-disordered boundaries are shown in stick representation. The non-superposed PTP, WPD and E loops of HePTP0, HePTP24 and HePTP240 are shown in (b), (c) and (d), respectively. (e) Residue Pro235 adopts the ‘atypically open’ conformation in HePTP240. Superposition of the WPD loops from HePTP0 (blue) and HePTP240 (red). In the WPD-closed structure (i.e. HePTP0; blue), residue Pro240 packs into the hydrophobic pocket formed by Phe231, Trp234, Leu246, and Phe321. In the WPD-open structure (i.e. HePTP240; red), residue Pro240 is disordered (illustrated as dashed lines), and residue Pro235 is rotated upward by ~90° (illustrated as a gradated, curved arrow) and instead packs into the Phe231/Trp234/Leu246/Phe321 pocket. The conformation of the conserved WPD loop proline (i.e. Pro235 in HePTP) in HePTP240 is identical to that observed in ‘atypically open’ structures of STEP,9 Lyp29 and GLEPP16;24. Protein models prepared using PyMOL.36