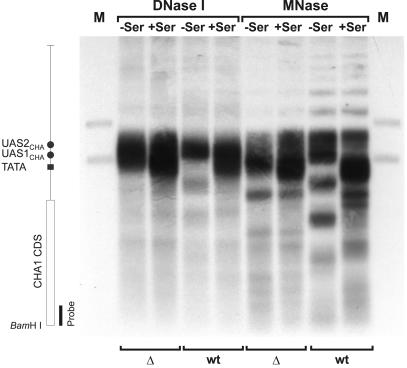
Fig. 3. Chromatin analysis of the CHA1 gene in the Δnhp6a Δnhp6b double mutant (RJY6012; Δ) and the isogenic wild-type strain (SEY6210; wt). MNase- and DNase I-based mapping of nucleosome organization was carried out as described previously (Moreira and Holmberg, 1998). Cells were grown in the absence (–Ser) or presence (+Ser) of serine, nuclei purified and digested for 10 min with 20 U/ml DNase I or 100 U/ml MNase. DNA was isolated, digested with BamHI, separated on a 1% agarose gel, blotted and hybridized with a 32P-labeled CHA1-specific PCR amplificate. Lanes M contain restriction enzyme double digests of genomic DNA with BamHI and ClaI or BamHI and HindIII, to generate position marker fragments. The vertical map indicates the relative positions of the various cis-acting sequences and the CHA1 coding sequence (CDS).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.