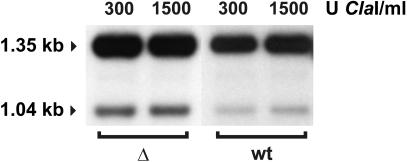
Fig. 4. Chromatin accessibility analysis of the PHO5 promoter in cells lacking NHP6A and NHP6B. Restriction enzyme-based mapping of nucleosome organization was carried out essentially as described in the legend for Figure 3, except that isolated nuclei were digested for 30 min with ClaI. DNA was isolated, digested with ApaI, separated on a 1% agarose gel, blotted, and hybridized with a 32P-labeled PHO5-specific PCR amplificate. A 1.35 kb ApaI fragment is generated in the absence of cleavage by ClaI, and a 1.04 kb fragment is generated if the ClaI site is accessible.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.