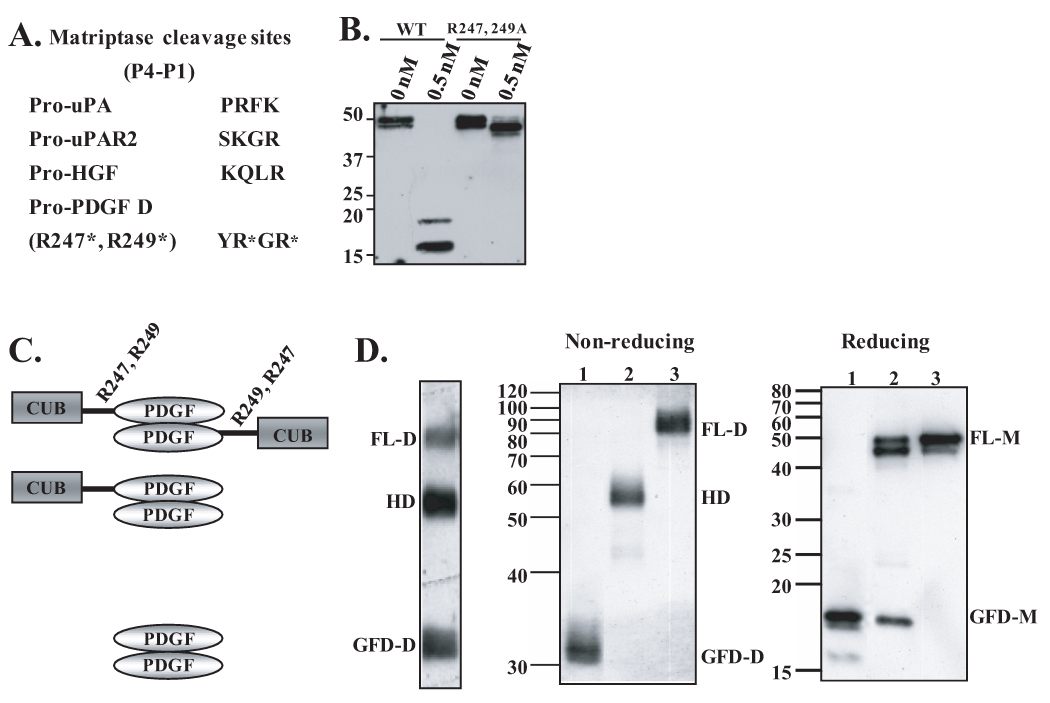
Fig. 3. Identification of matriptase cleavage site in the hinge region between the CUB and growth factor domains and characterization of PDGF-D dimer species.
A. Comparison of cleavage sites of known matriptase substrates, pro-uPA, pro-uPAR2, and pro-HGF, to the putative matriptase cleavage site within hinge region of full-length PDGF-D. B. Wild-type rPDGF-D and rPDGF-DR247,249A mutant proteins, in which arginine residues at amino acid 247 and 249 are mutated to alanines (6), were incubated with 0.5 nM matriptase overnight at 37°C. Samples were analyzed by immunoblot analysis in reducing condition. C. Schematic diagram of FL-D, HD, and GFD-D. R247 and R249 in the hinge between the CUB and GFD are labeled. D. PDGF-D dimer species (FL-D, HD, and GFD-D) produced by HeLa cells were isolated from a non-reducing SDS-PAGE gel (left panel), resolved on non-reducing or reducing SDS-PAGE gel, then immunoblotted with anti-PDGF-D antibody.