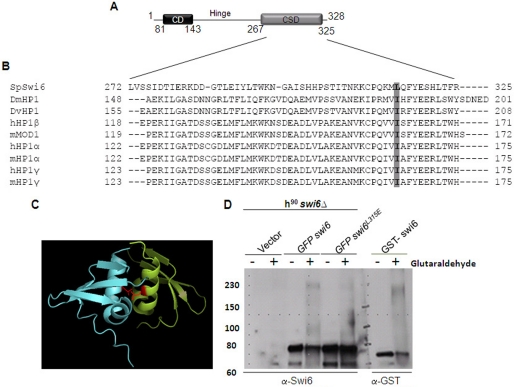
FIGURE 1.
L315E mutation in the CSD domain of Swi6 interferes with its self-association. A, schematic showing the domain structure of fission yeast Swi6. B, alignment of sequences of CSDs from different HP1 proteins numbered with respect to MOD1 (adapted from Ref. 7). The hydrophobic residue Leu-315 in Swi6 and Ile in other homologues is conserved and indicated by shading. C, a dimer formed by the association of the CSDs of two monomers. The monomers are shown in different colors. Two Leu-315 residues are shown in magenta in both CSD chains. Their side chains, shown as lines, are believed to play an important role in the association of two CSD monomers of Swi6 (5) (PDB code 1E0B). D, whole cell extracts of the swi6Δ mutant strain harboring empty vector (first and second lanes), GFP-swi6+ (3rd and 4th lanes), and swi6L315E mutant (5th and 6th lanes) genes expressed under the control of the nmt1 promoter and E. coli cells expressing GST-Swi6 (7th and 8th lane) were subjected to glutaraldehyde cross-linking and Western blotted with anti-Swi6 antibody. The GFP-tagged Swi6 monomer migrates at 78 kDa (≈ 28 (GFP) + 50 (Swi6)). Upon cross-linking, the multimer migrates around ∼230–240 kDa (A and B, 4th lane). The level of self-association is reduced in case of Swi6L315E mutant protein (A and B, 6th lane).