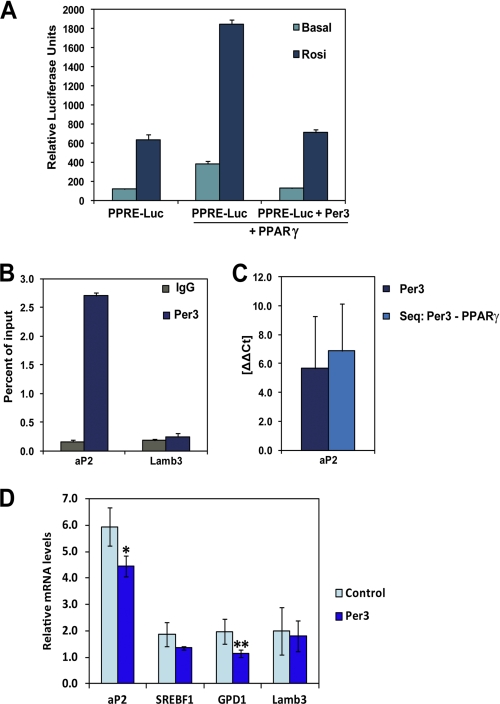
FIGURE 4.
Per3 inhibits PPARγ activity. A, COS-7 cells were transfected with a luciferase reporter gene containing three tandem PPARγ DNA binding motifs (PPRE-luc) with and without 100 nm rosiglitazone. Co-transfection with Pparγ induced reporter gene expression. The induction of reporter activity was significantly inhibited by the addition of Per3. Luciferase activity was internally normalized against the measured activity of a co-transfected Renilla expression vector. Three independent experiments were performed. Error bars represent S.D. between experiments. B, ChIP-qPCR of PER3 revealed enrichment near an endogenous PPRE in the enhancer region located −5.4 kb from the aP2 transcriptional start site compared with a control ChIP-qPCR of this region using a nonspecific IgG antibody. There was no significant enrichment of PER3 at another PPARγ binding site located within the Lamb3 gene (+7.6 kb from the transcriptional start site). The Per3 ChIP was performed three times, and error bars represent S.D. C, single ChIP of Per3 and sequential ChIP of Per3 followed by PPARγ was performed, and relative enrichment at the PPRE in aP2 was compared with the PPARγ binding site in Lamb3. After the sequential ChIP, qPCR of the Lamb3 site approximated the water control, whereas the aP2 site had significant enrichment indicating selective co-occupancy. D, transcript levels of PPARγ target genes in 3T3-L1 cells were measured with qPCR after 24 h of treatment with 100 nm rosiglitazone. Overexpression of Per3 resulted in significant decreased aP2 and Gpd1 transcript levels. Results were internally normalized to β-actin levels within each sample. Between three and five experiments were performed for each condition, and error bars represent S.E. *, p = 0.01; **, p = 0.03.