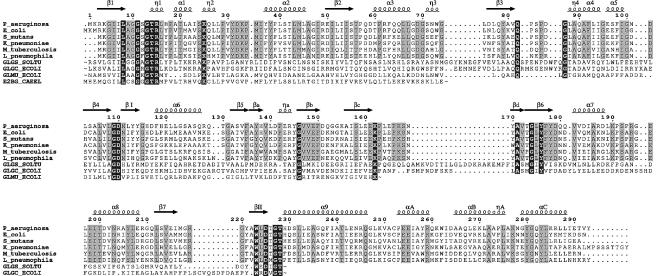
Fig. 3. Sequence alignment, prepared with CLUSTAL_X (Thompson et al., 1997), and manually edited to avoid deletions and insertions in secondary structure elements. Non-RmlA sequences were truncated as indicated by ‘∼’ in clearly unrelated regions. White letters on a black background represent conservation throughout all sequences while conservation within RmlA proteins only is shown by black letters on grey background. The RmlA sequences are as follows: P_aeruginosa, P.aeruginosa; E_coli, E.coli; S_mutans, Streptococcus mutans; K_pneumoniae, Klebsiella pneumoniae; M_tuberculosis, Mycobacterium tuberculosis; L_pneumophila, Legionella pneumophilia. Other sequences are: GLGS_SOLTU, Solanum tuberosum G-1-P adenylyltransferase small subunit (precursor); GLGC_ECOLI, E.coli G-1-P adenylyltransferase; GLMU_ECOLI, E.coli N-acetylglucosamine 1-phosphate uridyltransferase; E2BG_CAEEL, Caenorhabditis elegans putative translation initiation factor eIF2B γ-subunit. The figure was prepared with EsPript (Gouet et al., 1999).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.