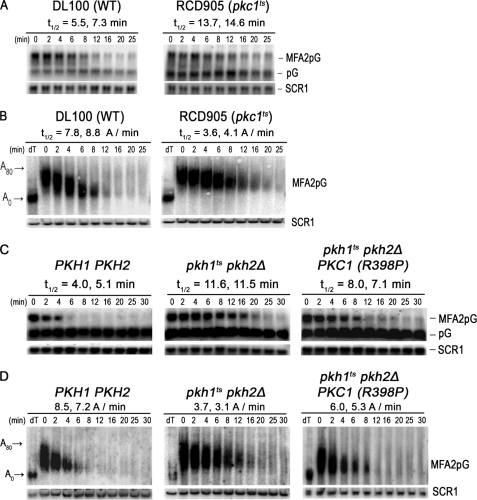
FIGURE 3.
Pkc1 controls mRNA decay and deadenylation. A, to determine whether Pkc1 controls MFA2pG mRNA decay, the rate of decay was examined in cells with wild-type PKC1 (DL100/pRP485-2) or a pkc1ts allele (RCD905/pRP485-2). Cells were grown at 39 °C for 60 min before transcriptional shut-off using glucose (4%) and thiolutin (4 μg/ml). RNA was extracted at the indicated time and analyzed on agarose gels. B, a portion of the RNAs shown in panel A was subjected to PAGE to examine the rate of deadenylation. RNA analyses were done as described in the legend to Figs. 1 and 2. C, to demonstrate that Pkc1 is the downstream effector of Pkh1/2 responsible for regulating mRNA decay and deadenylation, we compared MFA2pG mRNA decay in pkhts cells (RCD856/pRP485/pRS315-PKC1R398P) transformed with a plasmid expressing an allele of PKC1 encoding a constitutively active kinase to WT (Y7092/pRP485/pRS315) and pkhts cells transformed with just the vector (RCD856/pRP485/pRS315). The procedures for growing cells, incubating them for 1 h at the restrictive temperature, and analyzing mRNA decay were done exactly as described in the legend for Fig. 1B. D, portions of the RNA samples used for the Northern blots shown in panel C were also examined by PAGE to measure rates of deadenylation. Lanes labeled dT show the mobility of mRNA without a poly(A) as indicated at the left of the figure (A0) or the position of an mRNA with an intact poly(A) tail (A80).