FIGURE 2.
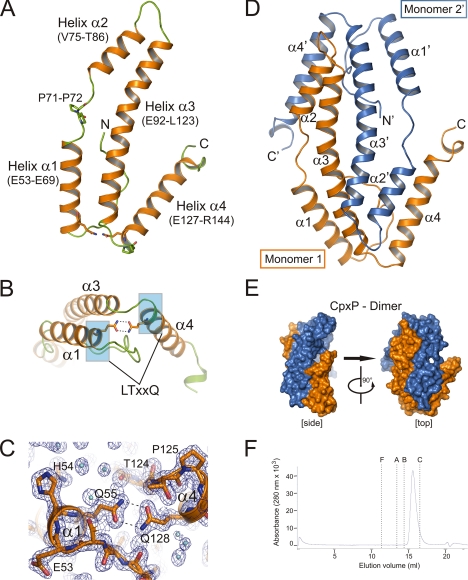
Crystal structure of CpxPΔ151. A, ribbon representation of a CpxPΔ151 monomer containing four helices α1 to α4 (helices, orange; loops, green). Two characteristic glutamine residues of the conserved LTXXQ repeat motifs (Leu51–Gln55 and Leu124–Gln128) and a PP motif (Pro71–Pro72) are shown as sticks. B, the monomer conformation with a V-shaped structure formed by helices α3 and α4 is stabilized by a double hydrogen bond between the conserved LTXXQ motifs (highlighted in blue) in helices α1 and α4. C, close-up view of the region surrounding the double hydrogen bond between Gln55 and Gln128 shown with 2Fo − Fc as electron density map (blue mesh) contoured at 1.2σ. D, the asymmetric unit contains a CpxPΔ151 dimer with two monomers that are interdigitated like left hands. Distinct perspectives of the CpxPΔ151 dimer are shown as surface representation (CpxPΔ151 monomer 1, blue; CpxPΔ151 monomer 2′, orange). E, ribbon representation of the CpxPΔ151 dimer using the same color scheme as in D. F, size-exclusion chromatography of CpxP was carried out on a Superdex 200 (HR10/30) column (GE Healthcare) equilibrated in 50 mm Tris/HCl, pH 7.5, containing 150 mm NaCl and 0.5% glycerol (v/v) at 4 °C. Protein samples (100 μl at 0.7 mg ml−1) were applied to the column at a flow rate of 0.5 ml min−1. The elution volumes for the molecular weight standards are shown by dashed lines (F = ferritin, A = aldolase, B = bovine serum albumin, C = carbonic anhydrase). See supplemental Fig. S3 for molecular mass determination of CpxPΔ151 by the combination of size-exclusion chromatography and sucrose-gradient sedimentation analysis.