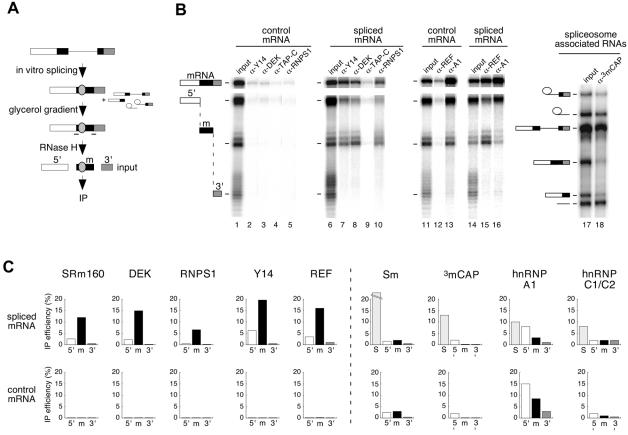
Fig. 3. Identification of proteins associated with the exon–exon junction region by RNA co-IP. (A) Scheme of co-IP strategy. AdML pre-mRNA was spliced for 2 h, and mRNP was separated from spliceosomes by glycerol gradient fractionation. Spliced mRNA was subsequently cleaved using RNase H and two oligos (–) to produce three fragments: 5′ (white), m (black) and 3′ (shaded). The fragment ‘m’ contains the –20/24 protected region (gray oval). This reaction mixture was then subjected to IP with different antibodies. Control mRNA was analyzed in parallel reactions except that it was not fractionated on a glycerol gradient. (B) Representative RNA co-IP experiments performed with antibodies against Y14, DEK, TAP-C, RNPS1, REF, hnRNPA1 (A1) and 3mCAP. Lanes 1, 6, 11, 14 and 17 correspond to one-fifteenth of input RNA. Lane 18 corresponds to co-IP of spliceosome-associated RNAs. AdML RNAs were analyzed as indicated in Figure 1. Structures of splicing substrates, intermediates and products as well as mRNA fragments are diagrammed. (C) Co-IP efficiencies of mRNA fragments (5′, m and 3′) and spliceosomes (S, taken as splicing intermediates) with antibodies against the species indicated. Upper and lower rows of histograms correspond to fragments of spliced and control mRNAs, respectively. Co-IP efficiencies are averages of 2–5 independent experiments.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.