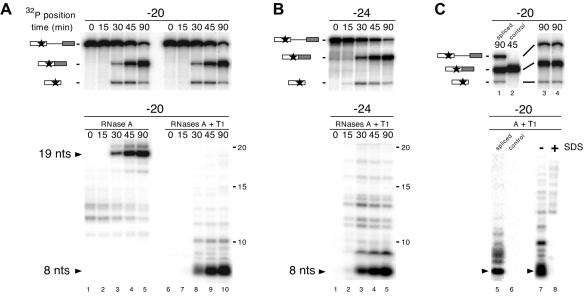
Fig. 4. Complete RNase digestion of singly labeled RNAs. (A) Upper panel: splicing time courses for AdML pre-mRNAs containing a single labeled phosphate (star) positioned 20 nt upstream of the 5′ splice site. Splicing substrates, intermediates and products are indicated on the left. RNAs were analyzed as in Figure 1. Lower panel: complete RNase A (left) or RNase A + T1 (right) digestion of RNAs at each time point yielded 19 and 8 nt protected fragments (arrows). Protected fragments were separated by 20% denaturing PAGE. (B) Same as (A) with AdML pre-mRNAs containing a single labeled phosphate positioned 24 nt upstream of the 5′ splice site. (C) Upper panels: AdML pre-mRNA (lanes 1, 3 and 4) or control mRNA (lane 2) containing a single labeled phosphate positioned 20 nt upstream of the 5′ splice site was incubated under splicing conditions for the time indicated and separated by 20% (lanes 1 and 2) or 15% (lanes 3 and 4) denaturing PAGE. Lower panels: aliquots of each splicing reaction were subjected to complete RNase A + T1 digestion with (lane 8) or without (lanes 5–7) prior SDS treatment. Protected fragments were separated as in (A).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.