Abstract
Background
Cytomegalovirus UL54 DNA polymerase mutations observed in clinical specimens are of diagnostic significance if confirmed to affect antiviral drug susceptibility.
Objectives
Validate an updated recombinant phenotyping method to determine the degree of drug resistance conferred by previously uncharacterized UL54 sequence variants, in comparison with known resistance-related mutations.
Study Design
Bacterial artificial chromosome clones of viral DNA were mutagenized by recombination, transfected to produce live virus and phenotyped by standardized reporter-based yield reduction assays.
Results
Sixteen recombinant viruses were constructed, representing baseline sequences, known resistance-related mutations and amino acid changes of unproven significance from clinical specimens. Phenotypes of baseline strains and known mutants were comparable to results from prior methods and helped to resolve some published inconsistencies. Mutations F412L, F412S, L545W were newly confirmed to confer ganciclovir and cidofovir resistance, while Q578H conferred ganciclovir and foscarnet resistance with borderline cidofovir resistance. Some foscarnet-resistant mutants were appreciably growth-retarded.
Conclusions
Results add to known exonuclease domain mutations that confer ganciclovir-cidofovir cross-resistance, polymerase domain mutations that confer foscarnet resistance with variably decreased ganciclovir/cidofovir susceptibility, and increase the list of sequence variants with no measurable impact on drug susceptibility. The phenotypic diversity of similar UL54 genotypic variants complicates the interpretation of genotypic resistance testing. Technical improvements are facilitating the phenotyping of remaining unknown sequence variants.
1. Background
Drug-resistant cytomegalovirus (CMV) is an ongoing concern especially in transplant recipients who are routinely exposed to extended prophylactic or treatment courses of ganciclovir (GCV), its oral prodrug valganciclovir, foscarnet (FOS) and/or cidofovir (CDV).1 Resistance testing is prompted by persistent plasma viral loads or overt disease after prolonged therapy. Because direct susceptibility testing of clinical CMV culture isolates is impractically slow, genotypic testing is used instead to detect viral drug resistance mutations in clinical specimens. Accurate interpretation of such testing depends on a validated database of genotype-phenotype correlations that is substantial but incomplete for CMV.2
Most GCV-resistant CMV isolates contain one of the canonical mutations in the viral UL97 kinase gene involved in the phosphorylation of GCV.2 Less commonly, mutations in the viral UL54 DNA polymerase (pol) gene may confer resistance to one or more of the current drugs. Larger3, 4 and smaller2 studies indicate that pol mutations conferring GCV and CDV resistance are concentrated in the exonuclease domains (codons 301, 408–413, 501–545) and region V (codons 981–987), while FOS resistance mutations range more widely, clustering in and between regions II and III (codons 696–845). Some early studies did not find any FOS-GCV cross-resistance,4 but later reports suggest that FOS resistance mutations may variably decrease susceptibility to GCV and CDV.2 Interpretation of pol sequence variants is further complicated by considerable baseline sequence polymorphism.2
The role of individual mutations in drug resistance is evaluated by their transfer to a control laboratory CMV strain and determining the resulting effect on drug susceptibility. This recombinant phenotyping process has historically been complicated by low operational efficiency.2 A recently developed CMV bacterial artificial chromosome (BAC) clone containing a reporter gene facilitated the phenotyping of many UL97 gene variants5 and was adapted here for phenotyping pol variants.
2. Objectives
(1) To validate a new BAC-based pol recombinant phenotyping method by comparison of results with previously published findings on known baseline and mutant strains; (2) Newly define the phenotypes of uncharacterized pol mutations plausibly linked to drug resistance that were found in CMV sequences from diagnostic laboratories.
3. Study Design
3.1 Viral strains and clones
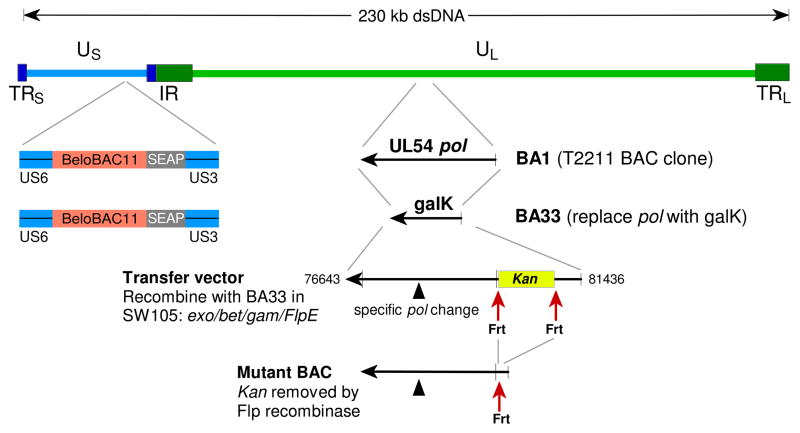
CMV strain T2211 was derived from standard strain AD169 and features a secreted alkaline phophatase (SEAP) reporter gene for rapid viral quantitation.6 It was cloned as BAC BA15 and used to derive BAC BA33, where the entire pol coding sequence was replaced with a bacterial galK expression cassette by recombineering (Fig. 1),7 as described for the UL97 region galK clone BA9.5 BA33 was propagated in E. coli host strain SW105.5, 7
Figure 1. Construction of mutant BACs.
The entire UL54 DNA polymerase sequence of the SEAP reporter strain T2211 BAC clone BA1 was replaced with a bacterial galK cassette, forming clone BA33. The desired pol sequence configuration was then introduced by recombination of a transfer vector into BA33 and positive selection for the Kan marker, which was subsequently removed by an induced Flp recombinase. The recombinant BACs were transfected into fibroblast cultures to reconstitute live CMV.
3.2 Transfer vectors
Strain AD169 pol region nucleotides 76643 through 81436 (Genbank sequence X17403) were cloned into Bluescript vector pBS2KS+ (Stratagene) using restriction sites SacI and EcoRI. The clone was mutagenized to create a Bsu36I restriction site 23–29 nucleotides upstream of the start of the pol coding sequence, with removal of 34 nucleotides of native AD169 sequence upstream of the created restriction site. A Bsu36I-Frt-flanked kanamycin resistance cassette (Kan), the same as used for vector SC408,5 was ligated into the Bsu36I restriction site, creating clone SC409. Derivatives of SC409, created by replacing the BglII-BspE1 restriction fragment (pol codons 324–1107) with products of PCR mutagenesis, were used as transfer vectors to introduce the desired pol sequence configurations into BAC clone BA33, as previously described for UL97 mutations.5 Following arabinose-induced Flp recombinase-mediated removal of the bacterial Kan cassette, a final BAC clone was obtained (Fig. 1) which contained a 34-nucleotide Frt motif upstream of pol. All BAC clones were checked for an intact HindIII digest pattern.5 Because of distinctive restriction digest pattern changes, it was possible to verify the success of each construction step (Fig. 2).
Figure 2. Restriction digest patterns of BAC DNA.
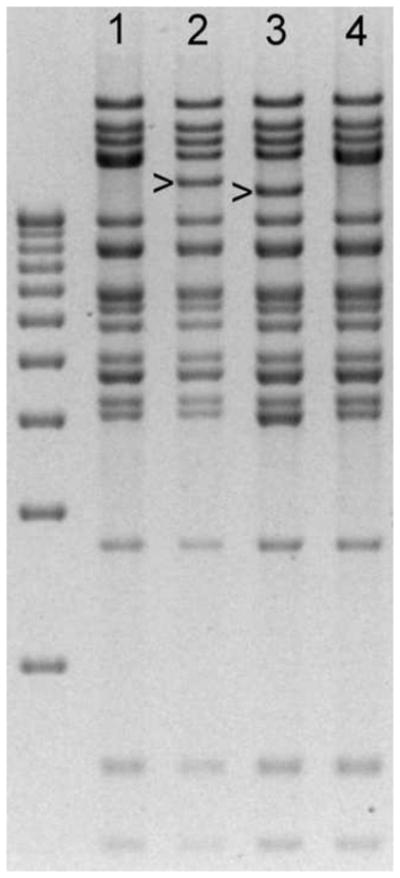
BAC DNAs were digested with enzyme HindIII and run on a field inversion agarose gel.5 Lane 1: BA1; Lane 2: BA33; Lane 3: BA33 after recombination with transfer vector containing the Frt-Kan cassette; Lane 4: Final recombinant BAC after removal of Kan marker, restoring the original digest pattern of BA1 (lane 1). Bands distinctive to each BAC construct are indicated. Sequential changes in restriction digest patterns were used to monitor the progress of each recombinant virus construction. The left lane is a ladder of DNA size markers ranging from 3kb to 12kb at 1-kb intervals.
3.3 Phenotypic assays
BAC DNA was transfected into human foreskin fibroblast (HFF) to reconstitute and propagate live CMV.5 Normal-appearing cytopathic effect was observed for all the mutants reported here. Recombinant strains were checked throughout pol for the intended sequence. Cell-free virus stocks were prepared and phenotypic assays were performed by SEAP reporter yield reduction as standardized in previous reports.2, 5, 6, 8 Briefly, a calibrated low multiplicity inoculum of cell-free virus stock was inoculated into 6 wells of a 24-well HFF cultures and incubated for 6 days under serial drug dilutions to determine the drug concentration required to reduce culture supernatant SEAP activity by 50% (EC50) as measured using a chemiluminescent substrate.6 Two replicate assays were performed at a time, and only assays meeting defined criteria pertaining to viral inoculum, adequate viral growth at 6 days, proper curve fit of SEAP values under different drug concentrations, and valid results with known controls5, 6 were accepted for reporting. Susceptibility phenotypes were assigned based on the mean EC50 values from assays set up on at least 4 different dates, to allow for variation in cell culture conditions. An EC50 value of 2-fold or greater than that of a matching susceptible control strain was used as the conventional criterion for drug resistance.2, 4, 5
3.4 Growth assays
Multicycle growth assays were performed as previously described6, 8–10 based on measurement of supernatant SEAP activities, with the initial low multiplicity viral inoculum calibrated among strains being compared, by measurement of SEAP activity at 24 hours. Comparisons among strains were set up in parallel using 3 to 4 replicate wells per strain.
3.5 Sequence variants selected for phenotyping
Control baseline BAC clones were created to represent the upstream Frt motif, and common pol amino acid polymorphisms S655L, N685S, A885T and N898D as found in Towne strain (Genbank HS5Towne). Representative drug resistance mutations were constructed and tested as BAC clones to compare with prior results.2 These included exonuclease domain mutations N408K and P522S, mutations in the pol catalytic domains (D588N, E756K, V781I, A809V and A987G). Mutations found in clinical isolates and suspected to confer drug resistance but not specifically validated by recombinant phenotyping included F412L 11, F412S 12, L545W and Q578H.13 Other pol sequence variants of undetermined significance and found after genotyping of specimens from treated individuals included C304S,14 V544A and A714V. These were chosen for phenotyping because of their close proximity to known resistance mutations D301N, L545S and V715M.2
4. Results
Table 1 shows the GCV, FOS and CDV susceptibilities of baseline and variant CMV strains as assessed by their mean EC50 values when compared to a matching baseline control strain.
Table 1.
Genotypes and Phenotypes of Recombinant Viruses
| BAC | Virus | UL54 pol genotype | Ganciclovir |
Foscarnet |
Cidofovir |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation1 | Baseline2 | EC503 | SD4 | N5 | Ratio6 | EC503 | SD4 | N5 | Ratio6 | EC503 | SD4 | N5 | Ratio6 | ||
| Baseline Strains | |||||||||||||||
| 2211 | S897L | 1.06 | 0.27 | 71 | 33 | 9 | 42 | 0.18 | 0.05 | 41 | |||||
| BA31 | 3265 | Frt, S897L | 1.18 | 0.38 | 54 | 40 | 14 | 39 | 0.18 | 0.06 | 33 | ||||
| BA104 | 3413 | Frt, S655L, N685S, A885T, N898D | 1.23 | 0.35 | 14 | 42 | 17 | 9 | 0.19 | 0.04 | 8 | ||||
| Control Resistant Mutants | |||||||||||||||
| BA57 | 3313 | N408K | Frt, S897L | 4.04 | 1.60 | 36 | 3.4 | 32 | 7 | 9 | 0.8 | 3.83 | 0.57 | 8 | 21 |
| BA102 | 3412 | P522S | Frt, S897L | 2.83 | 0.89 | 14 | 2.4 | 49 | 16 | 9 | 1.2 | 0.43 | 0.18 | 15 | 2.4 |
| BA103 | 3408 | D588N | Frt, S897L | 2.36 | 0.57 | 13 | 2.0 | 112 | 33 | 12 | 2.8 | 0.24 | 0.08 | 13 | 1.3 |
| BA116 | 3430 | E756K | Frt, S897L | 2.21 | 0.63 | 11 | 1.9 | 141 | 42 | 12 | 3.5 | 0.31 | 0.06 | 12 | 1.7 |
| BA108 | 3417 | V781I | Frt, S897L | 2.86 | 0.76 | 13 | 2.4 | 156 | 41 | 10 | 3.9 | 0.27 | 0.09 | 11 | 1.5 |
| BA37 | 3271 | A809V | Frt | 2.36 | 0.75 | 25 | 2.0 | 160 | 51 | 23 | 4.0 | 0.29 | 0.09 | 14 | 1.6 |
| BA115 | 3429 | A987G | Frt, A885T, N898D | 7.30 | 2.28 | 9 | 6.2 | 35 | 9 | 11 | 0.9 | 0.95 | 0.34 | 21 | 5.3 |
| Newly Characterized Recombinant Phenotypes | |||||||||||||||
| BA78 | 3365 | C304S | Frt, S897L | 1.59 | 0.46 | 11 | 1.3 | 44 | 15 | 12 | 1.1 | 0.25 | 0.12 | 11 | 1.4 |
| BA35 | 3267 | F412L | Frt, S897L | 5.37 | 1.44 | 13 | 4.6 | 42 | 18 | 10 | 1.1 | 1.70 | 0.6 | 12 | 9.4 |
| BA69 | 3334 | F412S | Frt, S897L | 6.25 | 2.85 | 14 | 5.3 | 34 | 13 | 9 | 0.8 | 2.35 | 1.01 | 12 | 13 |
| BA71 | 3336 | V544A | Frt, S897L | 1.16 | 0.31 | 12 | 1.0 | 37 | 10 | 10 | 0.9 | 0.19 | 0.07 | 11 | 1.1 |
| BA96 | 3400 | L545W | Frt, S897L | 5.79 | 1.90 | 12 | 4.9 | 53 | 13 | 11 | 1.3 | 1.13 | 0.49 | 11 | 6.3 |
| BA112 | 3426 | Q578H | Frt, S897L | 3.90 | 1.35 | 13 | 3.3 | 180 | 54 | 10 | 4.5 | 0.41 | 0.14 | 10 | 2.3 |
| BA70 | 3353 | A714V | Frt, S897L | 0.94 | 0.20 | 8 | 0.8 | 30 | 6 | 15 | 0.8 | 0.13 | 0.04 | 9 | 0.7 |
Items associated with drug resistance shown in bold.
BAC Bacterial artificial chromosome CMV clone name
Virus Serial number of recombinant CMV strain derived by transfection of BAC into fibroblasts
Frt Frt recognition site upstream of gene, left after removing Kan-R selection marker by Flp recombinase
Targeted amino acid change introduced into BAC by recombination
Other sequence variation from strain AD169
Mean drug concentration (µM) required to reduce SEAP growth by 50% at 6 days post-infection
Standard deviation of the EC50 values
Number of assays (performed over at least 4 separate dates)
Ratio of EC50 to baseline strain
4.1 Phenotypes of baseline BAC-derived strains
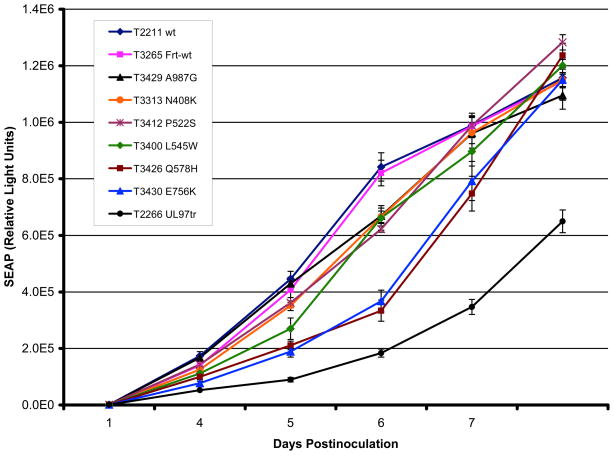
Strains T3265 and T3413 contain an upstream Frt motif and pol amino acid changes that are representative of interstrain polymorphism.2 The EC50s for GCV, FOS and CDV for T3265 and T3413 were not significantly different from the original strain T2211. Strain T3265 had a similar SEAP growth curve as its source strain T2211 (Fig. 3).
Figure 3. SEAP growth curves of baseline and mutant BACs.
Viral strains were inoculated at a multiplicity of infection of 0.02 as calibrated by supernatant SEAP activity at 24 hours. On each of days 4 to 8 postinoculation, culture supernatant was sampled for SEAP activity using a chemiluminescent substrate. Growth curves show the mean and standard deviation of values from 3 to 4 replicates of each strain set up in parallel. A severely growth inhibited UL97 kinase-defective mutant (T2266) was included as an additional control. The legend lists the strains in top to bottom order according to the position of their corresponding growth curves between days 5 and 6.
4.2 Phenotypes of known resistance-related mutations
Among pol mutations previously phenotyped without use of BAC clones,2 current results (Table 1) were consistent, with only minor differences in readouts of low-level cross-resistance. Mutations N408K, P522S and A987G have the same GCV-CDV dual resistance phenotypes as previously reported.4, 15 Mutations D588N, E756K, V781I and A809V that confer primarily FOS resistance (Table 1) are reported to have inconsistent or low levels of GCV or CDV resistance.2 The D588N mutant has shown low-grade GCV and CDV resistance,16 no GCV resistance,17 or a borderline GCV phenotype here. Similarly, an E756K mutant showed GCV and CDV EC50 ratios of 3.5 and 2.2,3 compared with 1.9 and 1.7 here, which are closer to ratios reported for another E756K mutant,18 and other mutants at codon 756.2 Mutation V781I was associated with GCV EC50 ratios ranging from 1.0 to 4.5, 4, 17 compared with 2.4 found here. Mutation A809V was previously associated with GCV and CDV EC50 ratios of 2.6 and 1.7,19 versus 2.0 and 1.6 here.
4.3 Phenotypes of uncharacterized sequence variants
Among pol mutations not previously validated by recombinant phenotyping (Table 1), F412L, F412S and L545W confer GCV resistance with substantial CDV cross-resistance, comparable to exonuclease mutants F412C, F412V and L545S.4, 20 Mutation Q578H was selected in vitro under FOS21 and also found in a clinical isolate.13 Current data (Table 1) show that it confers FOS resistance, with GCV and CDV cross-resistance. Of the 3 pol sequence variants C304S, V544A and A714V selected for their close proximity to known resistance mutations at codons 301, 545 and 715, none were found to confer significant drug resistance.
4.4 Growth properties
Comparison of multicycle growth curves by SEAP reporter assay (Fig. 3) showed perceptible growth retardation especially for the FOS-resistant mutants Q578H and E756K, consistent with prior reports of similar mutants 3, 9, 22. Exonuclease mutants, e.g. N408K, P522S and L545W, along with the region V mutation A987G, appeared to be less growth-attenuated than the FOS-resistant mutants.
5. Discussion
An updated recombinant phenotyping method for CMV pol mutations based on BAC-cloned control strains containing a SEAP reporter gene yielded similar results as previous studies done using more traditional techniques for known mutations. It newly validated some resistance mutations while not confirming a role in drug resistance for other pol sequence variants reasonably suspected to have one.
For the control resistance mutations in Table 1, findings confirm and help to interpret previous data that showed minor inconsistencies,2 including borderline or low levels of GCV cross-resistance for the FOS-resistance mutations D588N, E756K, V781I and A809V, at just above or below the EC50 ratio of 2 used as a cutoff for resistance. Such mutations can combine with UL97 mutations to increase the overall level of GCV resistance.9
Three of the newly confirmed GCV-CDV resistance mutations (F412L, F412S, L545W) involve different amino acid changes at the same codons (412 and 545) as previously implicated in drug resistance2. Codon 412 now has 4 different amino acid changes (F412C/V/L/S) known to confer a similar GCV-CDV phenotype, whereas at codon 522, amino acid change P522L did not confer the GCV-CDV resistance associated with P522A or P522S,23 indicating that mutations at the same codon cannot be assumed to have the same phenotype. Validation of Q578H as a FOS-resistance mutation with measurable GCV-CDV cross-resistance highlights the widening range of pol codons involved in FOS resistance. The codon range 492–588 is a region containing exonuclease III and polymerase delta region-C homologies among DNA polymerases2 and appears to be populated by both the typical exonuclease mutations conferring GCV-CDV dual resistance as well as FOS mutations (N495K, Q578H, D588N) with limited GCV or CDV cross-resistance.2
A number of pol sequence variants encountered in clinical specimens from treated subjects were considered plausibly linked to drug resistance because of their close proximity to proven resistance mutations, but C304S, V544A and A714V were not found to confer significant drug resistance. These variants add to an increasing list of pol amino acid changes that have been shown by recombinant phenotyping to confer no resistance to current drugs.18, 24 Their appearance in diagnostic specimens could reflect previously unrecognized baseline sequence polymorphism. Alternatively, the sequence changes may modulate drug susceptibility only in certain sequence contexts or combinations, or simply be artifacts of diagnostic PCR and sequencing.
BAC-based recombinant phenotyping technology5, 18 is advantageous, in part because the clonal character of BAC recombinants eliminates the need for CMV plaque purification in cell culture. The positive selection strategy chosen here (Fig. 1) for construction of BAC recombinants in E. coli, using a Kan cassette that was later removed to leave a residual 34-bp Frt motif that did not affect the drug resistance phenotype, is efficient and requires only a few clones to be screened for each recombinant construct (Fig. 2). An alternative counter-selection technique using deoxygalactose is available for introducing pol mutations using the same starting materials.7 This creates final recombinants which do not have an extraneous Frt motif, but requires considerably more screening effort to eliminate defective BACs.
Overall, recent technical improvements speed the work flow of assigning a drug susceptibility phenotype to uncharacterized pol amino acid changes, of which many remain2, 25 and require future analysis in order to refine the interpretation of genotypic resistance testing data.
Acknowledgments
I thank Gail Marousek, Victor Wong and Alwin Borgmann for excellent technical assistance, and Kirsten St. George for referring uncharacterized sequence variants for phenotyping. Reagents for recombineering were provided by the Biological Resources Branch of the National Cancer Institute. This work was supported by NIH grant AI39938 and Department of Veterans Affairs research funds.
Footnotes
CONFLICT OF INTEREST STATEMENT
The author has no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Snydman DR, et al. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation. 2010;89:779–85. doi: 10.1097/TP.0b013e3181cee42f. [DOI] [PubMed] [Google Scholar]
- 2.Lurain NS, Chou S. Antiviral drug resistance of human cytomegalovirus. Clin Microbiol Rev. 2010;23:689–712. doi: 10.1128/CMR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J Infect Dis. 2003;188:32–9. doi: 10.1086/375743. [DOI] [PubMed] [Google Scholar]
- 4.Cihlar T, Fuller MD, Cherrington JM. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–36. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou S. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob Agents Chemother. 2010;54:2371–8. doi: 10.1128/AAC.00186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou S, Van Wechel LC, Lichy HM, Marousek GI. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob Agents Chemother. 2005;49:2710–5. doi: 10.1128/AAC.49.7.2710-2715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou S. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob Agents Chemother. 2009;53:81–5. doi: 10.1128/AAC.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou S, Marousek GI, Van Wechel LC, Li S, Weinberg A. Growth and drug resistance phenotypes resulting from cytomegalovirus DNA polymerase region III mutations observed in clinical specimens. Antimicrob Agents Chemother. 2007;51:4160–2. doi: 10.1128/AAC.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou S, Van Wechel LC, Marousek GI. Cytomegalovirus UL97 kinase mutations that confer maribavir resistance. J Infect Dis. 2007;196:91–4. doi: 10.1086/518514. [DOI] [PubMed] [Google Scholar]
- 11.Humar A, Kumar D, Preiksaitis J, Boivin G, Siegal D, Fenton J, et al. A trial of valganciclovir prophylaxis for cytomegalovirus prevention in lung transplant recipients. Am J Transplant. 2005;5:1462–8. doi: 10.1111/j.1600-6143.2005.00866.x. [DOI] [PubMed] [Google Scholar]
- 12.Lurain NS, Bhorade SM, Pursell KJ, Avery RK, Yeldandi VV, Isada CM, et al. Analysis and characterization of antiviral drug-resistant cytomegalovirus isolates from solid organ transplant recipients. J Infect Dis. 2002;186:760–8. doi: 10.1086/342844. [DOI] [PubMed] [Google Scholar]
- 13.Oshima K, Kanda Y, Kako S, Asano-Mori Y, Watanabe T, Motokura T, et al. Case report: persistent cytomegalovirus (CMV) infection after haploidentical hematopoietic stem cell transplantation using in vivo alemtuzumab: emergence of resistant CMV due to mutations in the UL97 and UL54 genes. J Med Virol. 2008;80:1769–75. doi: 10.1002/jmv.21277. [DOI] [PubMed] [Google Scholar]
- 14.Cherrington JM, Fuller MD, Lamy PD, Miner R, Lalezari JP, Nuessle S, et al. In vitro antiviral susceptibilities of isolates from cytomegalovirus retinitis patients receiving first- or second-line cidofovir therapy: relationship to clinical outcome. J Infect Dis. 1998;178:1821–5. doi: 10.1086/314487. [DOI] [PubMed] [Google Scholar]
- 15.Scott GM, Weinberg A, Rawlinson WD, Chou S. Multidrug resistance conferred by novel DNA polymerase mutations in human cytomegalovirus isolates. Antimicrob Agents Chemother. 2007;51:89–94. doi: 10.1128/AAC.00633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Springer KL, Chou S, Li S, Giller RH, Quinones R, Shira JE, et al. How evolution of mutations conferring drug resistance affects viral dynamics and clinical outcomes of cytomegalovirus-infected hematopoietic cell transplant recipients. J Clin Microbiol. 2005;43:208–13. doi: 10.1128/JCM.43.1.208-213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mousavi-Jazi M, Schloss L, Drew WL, Linde A, Miner RC, Harmenberg J, et al. Variations in the cytomegalovirus DNA polymerase and phosphotransferase genes in relation to foscarnet and ganciclovir sensitivity. J Clin Virol. 2001;23:1–15. doi: 10.1016/s1386-6532(01)00160-3. [DOI] [PubMed] [Google Scholar]
- 18.Chevillotte M, Ersing I, Mertens T, von Einem J. Differentiation between polymorphisms and resistance associated mutations in the human cytomegalovirus DNA polymerase. Antimicrob Agents Chemother. 2010;54:5004–11. doi: 10.1128/AAC.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou S, Marousek G, Parenti DM, Gordon SM, LaVoy AG, Ross JG, et al. Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J Infect Dis. 1998;178:526–30. doi: 10.1086/515648. [DOI] [PubMed] [Google Scholar]
- 20.Chou S, Marousek G, Guentzel S, Follansbee SE, Poscher ME, Lalezari JP, et al. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–9. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 21.Mousavi-Jazi M, Schloss L, Wahren B, Brytting M. Point mutations induced by foscarnet (PFA) in the human cytomegalovirus DNA polymerase. J Clin Virol. 2003;26:301–6. doi: 10.1016/s1386-6532(02)00046-x. [DOI] [PubMed] [Google Scholar]
- 22.Baldanti F, Underwood MR, Stanat SC, Biron KK, Chou S, Sarasini A, et al. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J Virol. 1996;70:1390–5. doi: 10.1128/jvi.70.3.1390-1395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou S, Marousek G, Li S, Weinberg A. Contrasting drug resistance phenotypes resulting from cytomegalovirus DNA polymerase mutations at the same exonuclease locus. J Clin Virol. 2008;43:107–9. doi: 10.1016/j.jcv.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou S, Marousek G, Boivin G, Goyette N, Farhan M, Ives JAL, et al. Recombinant phenotyping of cytomegalovirus sequence variants detected after 200 or 100 days of valganciclovir prophylaxis. Transplantation. 2010;90:1409–13. doi: 10.1097/TP.0b013e3181fdd9d2. [DOI] [PubMed] [Google Scholar]
- 25.Boivin G, Goyette N, Rollag H, Jardine AG, Pescovitz MD, Asberg A, et al. Cytomegalovirus resistance in solid organ transplant recipients treated with intravenous ganciclovir or oral valganciclovir. Antivir Ther. 2009;14:697–704. [PubMed] [Google Scholar]