Figure 3.
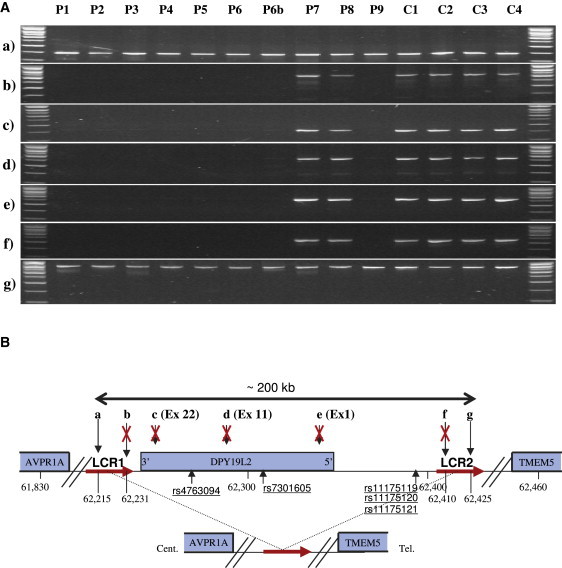
Characterization of a Large Homozygous Deletion Encompassing Only DPY19L2 in Eight of Ten Patients
(A) Seven loci (a–g) localized on and around DPY19L2 were amplified from ten patients with globozoospermia (P1–P9, including P6b) and four fertile controls (C1–C4). All of the tested loci yielded good PCR amplification from fertile individuals, whereas in eight of the ten globozoospermic patients there was no amplification of any of the intragenic loci (exons 1, 11, and 22) or of extragenic loci (b and f).
Genomic DNA was extracted either from peripheral blood leucocytes with the use of a guanidium chloride extraction procedure or from saliva via an Oragene DNA Self-Collection Kit (DNAgenotech, Ottawa, Canada). PCR primers with their specific annealing temperatures and exact genomic localization are listed in Table S1. Thirty-five cycles of PCR amplification were carried out with the use of Taq DNA polymerase (QIAGEN, Courtaboeuf, France).
(B) Schematic representation of the analyzed region with the position of the amplified loci (in kb). (a and b) Loci are localized approximately 25 kb and 9 kb 3′ of DPY19L2, respectively; (c), (d), and (e) show the position of DPY19L2 exons 22, 11, and 1, respectively; and (f) and (g) loci are localized approximately 62 and 77 kb 5′ of DPY19L2. LCRs 1 and 2 (red arrows) represent two 28 kb duplicated sequences showing 97% sequence identities located on each side of DPY19L2. PCR results in (A) indicate that the centromeric breakpoint is located between loci (a) and (b) (16 kb) within LCR 1 and the telomeric breakpoint between loci (f) and (g) (15 kb) within LCR 2. The deleted region encompasses a maximum of 210 kb, containing only one gene: DPY19L2.
