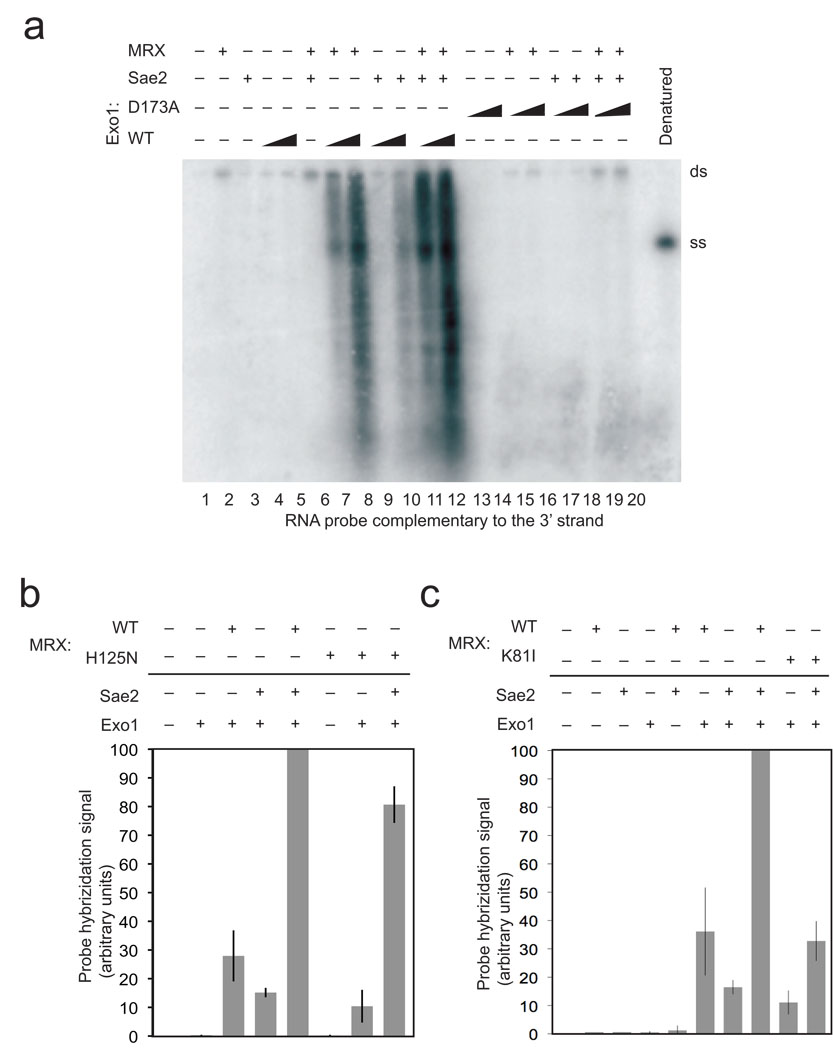
Figure 4. Mutations in Exo1 and Rad50 prevent DNA end processing.
(a) Resection assays were performed and analyzed as in Fig. 1B using non-denaturing southern hybridication with a 3' strand probe and with a catalytic mutant of Exo1, D173A, as indicated. Concentrations of wild-type and mutant Exo1 were 4 nM and 8 nM. (b) Resection assays were performed as in Fig. 1B with the probe specific for the 3' strand but with MRX complexes containing the Mre11 mutant H125N. Results from 3 experiments were quantitated using phosphorimager analysis to quantitate the total counts in each lane and the average of these shown, with error bars indicating standard deviation. Within each experiment, the signal from the reactions containing wild-type MRX, Exo1, and Sae2 was the highest and set to 100%, with the signals in other lanes shown relative to this value. (c) Resection assays were performed, analyzed, and presented as in (b) but with the MR(K81I)X mutant complex. Reactions from three independent experiments were quantitated.