Abstract
A major challenge for core facilities is determining quantitative protein differences across complex biological samples. Although there are numerous techniques in the literature for relative and absolute protein quantification, the majority is nonroutine and can be challenging to carry out effectively. There are few studies comparing these technologies in terms of their reproducibility, accuracy, and precision, and no studies to date deal with performance across multiple laboratories with varied levels of expertise. Here, we describe an Association of Biomolecular Resource Facilities (ABRF) Proteomics Research Group (PRG) study based on samples composed of a complex protein mixture into which 12 known proteins were added at varying but defined ratios. All of the proteins were present at the same concentration in each of three tubes that were provided. The primary goal of this study was to allow each laboratory to evaluate its capabilities and approaches with regard to: detection and identification of proteins spiked into samples that also contain complex mixtures of background proteins and determination of relative quantities of the spiked proteins. The results returned by 43 participants were compiled by the PRG, which also collected information about the strategies used to assess overall performance and as an aid to development of optimized protocols for the methodologies used. The most accurate results were generally reported by the most experienced laboratories. Among laboratories that used the same technique, values that were closer to the expected ratio were obtained by more experienced groups.
Keywords: Association of Biomolecular Resource Facilities study, protein quantification, protein quantitation
INTRODUCTION
There are numerous different methods to interrogate the proteome in a quantitative manner. In general, they fall into two major categories: methods based on two-dimensional (2-D) gel electrophoresis with poststaining1,2 or prelabeling3,4 or methods in which quantification is carried out using mass spectrometric measurement at the peptide level. Methods for relative quantification of peptides have been developed using stable isotope labeling in vitro5–7 and in vivo,8 as well as label-free approaches.9,10
A major challenge for proteomics laboratories is to determine differences in protein abundance among biological samples. Most of the applicable approaches are not routine, can be challenging to implement effectively, and are made more difficult by the complexity of the mixtures to which they are applied. There are few studies in which these technologies have been compared in terms of reproducibility, accuracy, and precision. Moreover, there are no studies to date dealing with analytical performance across multiple laboratories with varied levels of expertise.
In 2006, the Proteomics Research Group (PRG) of the Association of Biomolecular Resource Facilities (ABRF) began examining some of these variables in a study where participating laboratories received two samples that contained eight proteins in differing ratios.11 A variety of quantitative approaches were used by the participants. The 2006 study was the first of its kind to chart the breadth of technologies used by different laboratories, the variability in the accuracy of data returned, and the differing levels of expertise within proteomics facilities. However, the design of the study left many questions unanswered. In particular, the performance of the methods in the presence of a complex mixture of background proteins was not investigated.
The goal of the PRG 2007 study was to expand the 2006 study by focusing on identifying and quantifying proteins within a complex mixture. In addition to making it possible for participating laboratories to assess their own capabilities in this regard, the results of the study would permit the proteomics community in general to gain a relative measure of success of the different quantitative approaches used.
As a part of the study, the PRG also collected and compiled supplemental information about the strategies used by each participant; this was undertaken as a way to provide an overview of the protocols and techniques used and to aid in the development of optimized protocols for these techniques. Finally, information was collected with respect to length of time that the participants had been using the technologies they applied to this study sample to ascertain if there was a correlation between experience and successful use of a technology.
MATERIALS AND METHODS
Each sample set consisted of three tubes (labeled A, B, and C). Tubes B and C were identical, although this information was not provided to the participants. Each tube was prepared by combining approximately 100 μg of a complex protein mixture (Escherichia coli lysate) with 12 commercially available, known proteins (“spikes”) that were added in varying quantities. The total amount of spiked proteins was 1.4 μg/tube. The same amount of the background protein mixture was added to each of the three tubes, and then the samples were lyophilized. Each tube thus contained 101.4 μg of a mixture of lyophilized proteins in which the background proteins were present at the same relative concentrations in each tube, and the added spikes were present at different amounts. The identities, quantities, and ratios of added proteins are listed in Table 1.
TABLE 1.
Proteins in PRG07 study samples
| Proteinb | Accession numberc | M.W. (kDa) | Quantity (pmol)a |
Ratio (B/A) | |
|---|---|---|---|---|---|
| A | B | ||||
| Myoglobin | 161 | 16.5 | 0.50 | 5.00 | 10.00 |
| Ubiquitin | 4014 | 8.7 | 5.00 | 23.00 | 4.60 |
| Cytochrome c | 3870 | 13.0 | 2.50 | 11.50 | 4.60 |
| HRP | 2466 | 43.3 | 5.00 | 11.00 | 2.20 |
| Serum albumin | 1213 | 66.6 | 5.00 | 3.33 | 0.67 |
| Catalase | 465 | 57.5 | 0.50 | 0.34 | 0.67 |
| Carbonic anhydrase I | 69 | 28.9 | 2.50 | 1.14 | 0.45 |
| Lactoperoxidase | 2648 | 77.5 | 2.50 | 0.78 | 0.31 |
| Glucose oxidase | 152 | 80.0 | 0.50 | 0.33 | 0.67 |
| Glycerokinased | 904 | 54.0 | 2.50 | 0.78 | 0.31 |
| Hexokinase | 2938 | 50.0 | 0.50 | 0.16 | 0.31 |
| Tryptophanased | 2366 | 51.0 | 5.00 | 1.56 | 0.31 |
aSample C contained the same quantities of protein as sample B.
bProteins were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA).
cAccession number in PRG database.
dAdded E. coli proteins.
Several of the more abundant spiked proteins were present at the ∼0.2-μg level, and the lower abundance spiked proteins were ∼0.01 μg. In some cases, isoforms and contaminants were also present, as is often the case with “real-life” biological samples. The dried mixtures were prepared from aqueous solutions that also contained small amounts of salts. There was no evidence that the samples contained any appreciable quantities of interfering substances that contained primary amino groups and/or free thiols. Participants were told that the samples had been dissolved successfully in 50–100 mM ammonium bicarbonate with about 20% acetonitrile but that other solvents might work.
Replicate sample sets were provided by the authors when requested so that participants would have a way to assess the reproducibility of their results. The participating laboratories were asked to identify the proteins that were present at different relative levels in the samples and to determine their relative quantities in the three samples. Results were returned using an on-line questionnaire.
The E. coli lysate (EC11303, made from lyophilized E. coli cells) and all added proteins were purchased from Sigma-Aldrich Chemical Co. Stock solutions of the E. coli lysate and the individual protein samples were prepared at a concentration of 1 mg/mL in 5% acetic acid/10% acetonitrile in deionized water. Protein purity values supplied by the vendor were used in the concentration calculations. The E. coli stock solution was apportioned into two conical tubes (A, 25 mL; B/C, 50 mL), and appropriate volumes of the individual protein stock solutions were added to the two tubes to produce the study test mixtures. Aliquots (100 μL) of A and B/C were transferred to the corresponding polypropylene sample tubes labeled A, B, and C. The mixtures were dried in a vacuum centrifuge and stored at –80°C prior to mailing. The samples were mailed at room temperature and were accompanied by a letter giving a description of the sample, the aims of the study, and instructions for submission to the PRG for analysis.
The PRG provided an anonymous protein database containing the sequences of the added and background proteins as well as a number of decoy protein entries. In addition, the study database contained frequently encountered experimental contaminants that were identified by name. The anonymous protein database was available as a download and was also accessible on the ProteinProspector, Mascot, and X! Tandem websites.
Participants were requested to report the anonymous accession numbers for up to 15 proteins that were found to be present at differing relative amounts between samples A and B and between samples B and C, along with a measure of their ratios. As not all laboratories were able to complete all of the requested analyses before the submission deadline, participants were asked to report results for comparison of A versus B as a minimum. Participants were also asked to indicate how confident they felt about each result and to keep track of how many hours their group spent planning the study experiments, preparing the samples, performing the analyses, and analyzing the data. To maintain anonymity in the study, when completing the on-line questionnaire, each participant entered a self-chosen, five-digit identifier; association between identifiers and participants was known only to an “anonymizer”, who was not a member of the PRG and who did not disclose the associations to any of the members of the PRG.
RESULTS
Sample Requests
Samples were requested by 87 laboratories; 43 participants (22 ABRF members and 21 nonmembers) submitted datasets, corresponding to a 49% return rate. Surveys from eight of the respondents did not contain any quantitative data.
Methods Used
Mass spectrometry (MS) was used for protein identification for all samples. Gel-based and gel-free approaches were used for quantification. Table 2 summarizes the methods used by the participants.
TABLE 2.
Methods used for quantification
| Methoda | Number |
|---|---|
| Gel-based (28%) | |
| 2-D Coomassie | 1 |
| 2-D silver-stain | 1 |
| 2-D fluorescence | 3 |
| 2-D DIGE | 5 |
| MS/isotope (55%) | |
| iTRAQ | 16 |
| 16O/18O | 2 |
| ICAT | 1 |
| ICPL | 1 |
| Label-free (17%) | 6 |
aDIGE, Difference gel electrophoresis; iTRAQ, isobaric tags for relative and absolute quantitation; ICAT, isotope-coded affinity tag; ICPL, isotope-coded protein label.
Graphical illustrations of the reported results for eight of the spiked proteins are shown in Fig. 1 and Fig. 2. Results for hexokinase, tryptophanase, glycerokinase, and glucose oxidase are not shown, as few laboratories reported results for these proteins. Details for all responses are shown in Supplemental Table 1. The “% error of ratio”, a numerical assessment of how close a submitted result for the relative quantities of a protein in samples A and B was to the expected ratio, was calculated as follows:
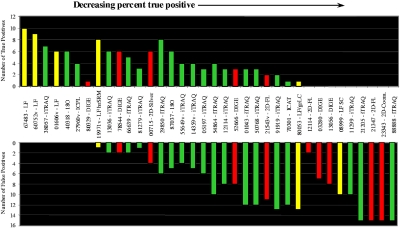
FIGURE 1.
PRG07 study results. The percent true-positive values decrease from left to right; percent true positives = [(number of true positives)/(number of true positives+number of false positives)] × 100; yellow, label-free (LF); green, MS-isotope; red, gel. The “number of true positives” is the number of proteins correctly identified as being present at different relative levels in samples A and B. The “number of false positives” is the number of proteins incorrectly identified as being present at different relative levels in samples A and B. The absolute number of proteins showing variation in expression level determined by each participating group is plotted, ranked in order of decreasing percentage of true-positive identification of the variation in the expression level. Those reporting only true positives appear first, followed by those reporting some true positives and some false positives, followed by those reporting only false positives.
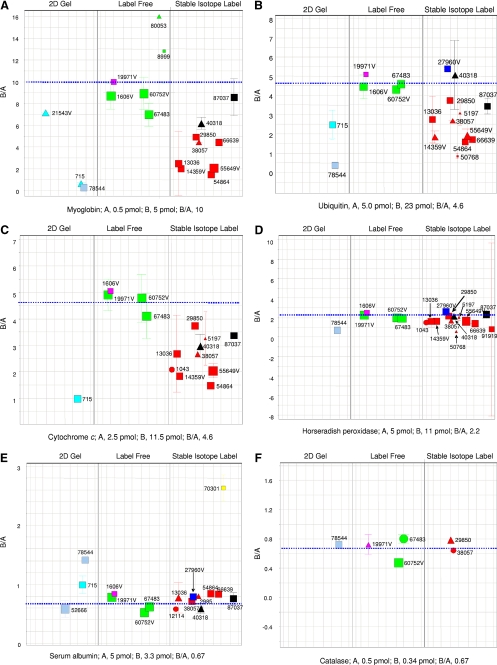
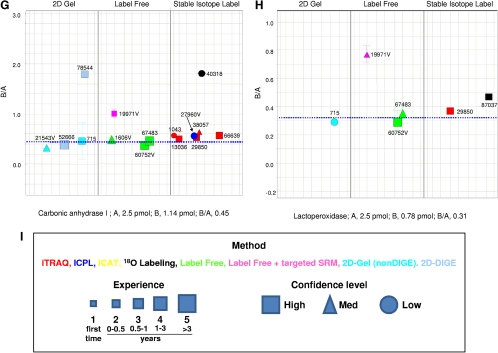
FIGURE 2.
Reported results for the individual spiked study proteins. The dashed blue line in each panel shows the expected mole ratio of B/A for the quantities listed below each panel. Each data point is coded for method (font color), experience (symbol size), and confidence level (symbol shape), as indicated in the box in this figure (last panel). Glucose oxidase, tryptophanase, glycerokinase, and hexokinase are not shown in this figure, as there were insufficient data for these proteins. The numbers shown adjacent to each symbol correspond to the anonymous identifiers chosen by the participating laboratories.
% error of ratio = [(observed ratio−expected ratio)/expected ratio] × 100
When examining these results, it is important to remember that the total number of participating laboratories was too small to draw statistically significant conclusions. In addition, many of the participants used these samples as a way to try out new methods that they had not attempted previously. Consequently, it is not reasonable to draw conclusions about the relative success of any specific method.
DISCUSSION
Results that were close to the expected values were reported by a few of the participants, indicating that quantitative assessment of complex samples is achievable. About one-third of the participants was able to identify and detect differences in the five most abundant proteins out of 10 added proteins. However, differences in the two added E. coli proteins were not detectable by any of the participants, most likely a result of the high endogenous levels of these proteins. In general, the most accurate results were reported by the most experienced laboratories. Among laboratories that used the same technique, values that were closer to the expected ratio were obtained by groups that had more experience with the technique. It is important to remember that direct comparisons of different approaches cannot be made on the basis of the results of this study because of the limited number of participants and the apparent dependence on experience. In addition, this was a model study in which biological variability was not a factor. In real-life samples, biological variability among samples would contribute substantially to the difficulty of the analysis. And finally, the only non-E. coli proteins in the PRG database (other than common contaminants) were the proteins that were spiked into the E. coli lysate. As such, it would have been possible to deduce the identity of these spikes by careful examination of the database. Taken together, the results of the PRG 2007 study indicate that successful quantitative proteomics requires a combination of appropriate instrumentation and experienced personnel.
For additional information, please visit http://www.abrf.org/PRG.
DISCLOSURES
The study and survey were undertaken with the goals of helping proteomics laboratories test, improve, and expand the range of their own capabilities. The PRG strongly points out that the data received from the study participants are not intended to promote any particular method or type of equipment. Furthermore, the number of submitted responses was insufficient to afford a statistically significant measure of the ability of any method to “get the correct answer”. The PRG also points out that in many cases, it is likely that the results represent the current experience levels of the scientists who performed the analyses and not the absolute capabilities of the methods used, as some of the participating laboratories were conducting these analyses for the first time. Any representation to the contrary of the above statements is the responsibility of the entity making that representation, and the PRG explicitly does not endorse any such representation.
ACKNOWLEDGMENTS
The PRG gratefully acknowledges the assistance of the following people: Kevin Hakala (The University of Texas Health Science Center at San Antonio, San Antonio, TX, USA), initial gel analyses; Michelle Salemi (University of California, Davis, CA, USA), sample management; Dr. Rich Eigenheer (University of California, Davis), HPLC-electrospray ionization (ESI)-MS/MS analyses; Ekaterina Deyanova (Merck, Rahway, NJ, USA), HPLC-ESI-MS/MS analyses; and Markus Hardt (University of California, San Francisco, CA, USA), anonymizer. The PRG also appreciates the hard work of the participants and their willingness to complete the results survey, even when no proteins were identified.
REFERENCES
- 1. Fievet J, Dillmann C, Lagniel D, et al. Assessing factors for reliable quantitative proteomics based on two-dimensional gel electrophoresis. Proteomics 2004;4:1939–1949 [DOI] [PubMed] [Google Scholar]
- 2. Smejkal GB, Robinson MH, Lazarev A. Comparison of fluorescent stains: relative photostability and differential staining of proteins in two-dimensional gels. Electrophoresis 2004;25:2511–2519 [DOI] [PubMed] [Google Scholar]
- 3. Yan JX, Devenish AT, Wait R, Stone T, Lewis S, Fowler S. Fluorescence two-dimensional difference gel electrophoresis and mass spectrometry based proteomic analysis of Escherichia coli. Proteomics 2002;2:1682–1698 [DOI] [PubMed] [Google Scholar]
- 4. Hu Y, Wang G, Chen GYJ, Fu X, Yao SQ. Proteome analysis of Saccharomyces cerevisiae under metal stress by two-dimensional differential gel electrophoresis. Electrophoresis 2003;24:1458–1470 [DOI] [PubMed] [Google Scholar]
- 5. Gygi SP, Rist R, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 1999;17:994–999 [DOI] [PubMed] [Google Scholar]
- 6. Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem 2001;73:2836–2842 [DOI] [PubMed] [Google Scholar]
- 7. Ross P, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154–1169 [DOI] [PubMed] [Google Scholar]
- 8. Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol Cell Proteomics 2004;3:729–735 [DOI] [PubMed] [Google Scholar]
- 9. Old WM, Meyer-Arendt K, Aveline-Wolf L, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 2005;4:1487–1502 [DOI] [PubMed] [Google Scholar]
- 10. Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res 2006;5:2909–2918 [DOI] [PubMed] [Google Scholar]
- 11. Turck CW, Falick AM, Kowalak JA, et al. ABRF-PRG2006 study: relative protein quantitation. Mol Cell Proteomics 2007;6:1291–1298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.