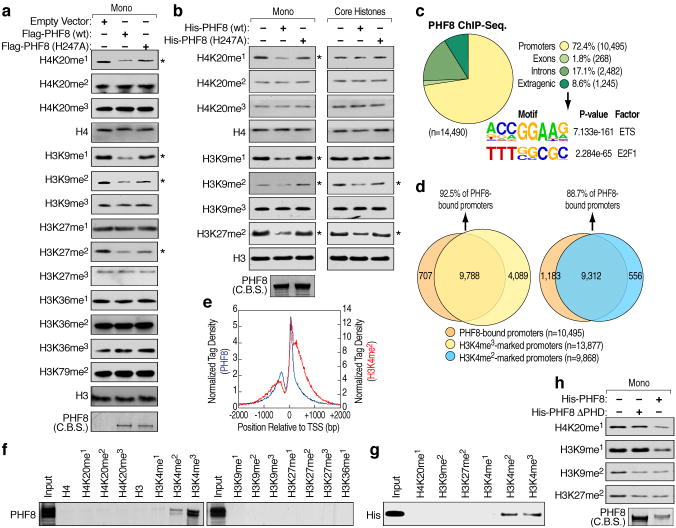
Figure 1. Histone demethylation mediated by PHF8.
a, Demethylase activity of Flag-tagged full-length wild type and mutant (H247A) PHF8 immunoprecipitated from HEK293T cell lysates was assessed using mononucleosomes as substrates. Expression of PHF8 proteins was visualized by Commassie blue staining (C.B.S). Asterisk denotes potential substrate. b, Demethylase activity of purified bacterially-expressed His-tagged full-length wild type and mutant (H247A) PHF8 proteins assessed using mononucleosomes (left panels) or core histones (right panels) as substrates. Expression of PHF8 proteins were visualized by Commassie blue staining (bottom left panel). Asterisk denotes potential substrate. c, Genomic distribution and top enriched motifs of PHF8 ChIP-seq. peaks (n=14,490) in HeLa cells. d, Venn diagrams showing overlap between PHF8-bound and H3K4me3 and H3K4me2-marked promoters. e, Tag density plots displaying PHF8 and H3K4me2 tags distribution relative to the transcriptional start site (TSS). f–g, Peptide pull-down assays mixing HeLa nuclear extracts (f) or purified bacterially-expressed His-tagged PHF8 PHD finger (aa1-54) (g) with biotinylated histone tails. Pull-downs were analyzed by immunoblotting. h, Demethylase activity of purified bacterially-expressed His-tagged wild type and ΔPHD finger (54–1024) PHF8 proteins assessed using mononucleosomes as substrates. Expression of PHF8 proteins were visualized by Commassie blue staining.