Abstract
We completed a biophysical characterization of the c-MYC proto-oncogene P1 promoter quadruplex and its interaction with a cationic porphyrin, 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin (TMPyP4), using differential scanning calorimetry, isothermal titration calorimetry, and circular dichroism spectroscopy. We examined three different 24-mer oligonucleotides, including the wild-type (WT) sequence found in the c-MYC P1 promoter and two mutant G→T sequences that are known to fold into single 1:2:1 and 1:6:1 loop isomer quadruplexes. Biophysical experiments were performed on all three oligonucleotide sequences at two different ionic strengths (30 mM [K+] and 130 mM [K+]). Differential scanning calorimetry experiments demonstrated that the WT quadruplex consists of a mixture of at least two different folded conformers at both ionic strengths, whereas both mutant sequences exhibit a single two-state melting transition at both ionic strengths. Isothermal titration calorimetry experiments demonstrated that both mutant sequences bind 4 mols of TMPyP4 to 1 mol of DNA, in similarity to the WT sequence. The circular dichroism spectroscopy signatures for all three oligonucleotides at both ionic strengths are consistent with an intramolecular parallel stranded G-quadruplex structure, and no change in quadruplex structure is observed upon addition of saturating amounts of TMPyP4 (i.e., 4:1 TMPyP4/DNA).
Introduction
It has been proposed that one of the most promising areas in cancer research is the design of drugs that focus on structural selectivity rather than sequence selectivity when targeting DNA or oncogene expression (1). G-quadruplex and i-Motif higher-order DNA structures have been the focus of recent strategies for oncogene regulation (1–5). Approximately 40% of all human genes have been found to have i-Motif-forming purine and G-quadruplex-forming pyrimidine-rich sequences within 1 Kb of their start sequences (1,6). Of the known human oncogenes, more than half have been shown to have G-quadruplex- and i-Motif-forming sequences in their promoter regions (7). One gene of particular interest for cancer treatment and possible prevention is the c-MYC oncogene, which is overexpressed in 60–80% of all cancers (1,2,5,8–11). Overexpression of the c-MYC oncogene can result in an increase of cellular proliferation and cell growth, and has been shown to influence and inhibit normal apoptosis (1,5,8–10). Some cancers associated with the overexpression of c-MYC include breast, colon, cervix, small cell lung, and myeloid leukemia (9,12,13). The examination of G-quadruplex structures found in the promoter region of the c-MYC oncogene and the intermolecular interactions between structure-specific small molecules and the higher-order DNA structure represents a new and promising area of cancer research (7,10,12,14,15).
In the attempt to design drugs for oncogene regulation, one of the most significant problems (and a subject of much debate) is the uncertainty as to whether the G-quadruplex and i-Motif higher-order structures are biologically relevant or exist at all under human physiological conditions (1,11,16,17). However, it has been established that binding a drug with high affinity for G-quadruplex drives the folding structural equilibrium in the direction of G-quadruplex formation (1,18). Several studies described the use of topoisomerase II inhibiters that interact with G-quadruplexes, and showed that TMPyP4 is able to further stabilize the G-quadruplex structure and suppress any further c-MYC transcriptional activation (10,12,14,19).
To date, biophysical studies have been limited to short single-stranded DNA sequences as models for the stability and structure of the G-quadruplex that can form in the promoter regions of several oncogenes. The model promoter region sequences have shown the potential to fold into multiple unique sequence conformations (1,8,9,12,20). The structural diversity and conformational equilibrium exhibited by these sequences are the source of the second most significant problem in designing drugs that target G-quadruplex (and/or i-Motif) structures, i.e., determining the most relevant target structure. To design a small molecule with very high affinity and selectivity for the c-MYC promoter region, we must first know the structure of each receptor G-quadruplex or at least the structure of the most significant target conformation. In previous studies, investigators attempted to determine the number and distribution of structural conformers simultaneously existing in dilute solution using multiple techniques (12,20,21). In this study, we determined the relative or equilibrium concentrations of two different folded isomers of the wild-type (WT) 24 mer c-MYC sequence by comparing the differential scanning calorimetry (DSC) profiles for the WT sequence and two mutant sequences that are known to fold into unique conformations. The biophysical characterization of the WT polypurine sequence found in the c-MYC NHE-IIII P1 promoter region, and two G→T mutant sequences were included in the determination of the thermodynamics of binding meso-tetra (N-methyl-4-pyridyl) porphine (TMPyP4). Both mutant constructs, as well as the WT, bind 4 mol of TMPyP4 at saturation, and there are at least two different binding modes for the interaction of the planar cationic porphyrin TMPyP4 with each of the c-MYC model G-quadruplexes (1,12,22–25).
Our data support the results of Phan et al. (8) in that we found that the WT purine-rich strand of the c-MYC proto-oncogene exists in dilute solution as an equilibrium mixture of multiple folded conformations. We also show that the affinity of TMPyP4 for binding to c-MYC promoter sequence G-quadruplexes is dependent on the folded conformation or structure (1,9). This work is significant because we were able to develop thermodynamic methods and models to describe the conformational equilibria in G-quadruplex-forming polypurine sequences.
Materials and Methods
In this study we used model c-MYC polypurine promoter oligonucleotides with either the native sequence (WT) or mutant sequences known to fold into the 1:2:1 and 1:6:1 loop isomers. Oligonucleotides were obtained from Oligos Etc. (Wilsonville, OR). The sequences for the WT and the mutant constructs are shown in Scheme 1. All sample handling, solution preparation, and experimental procedures used for the DSC, isothermal titration calorimetry (ITC), ultraviolet (UV), and circular dichroism (CD) measurements are described in detail in the Supporting Material.
Scheme 1.
c-MYC WT, 1:6:1 mutant, and 1:2:1 mutant P1 promoter sequences
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT 24 mer | 5′- | T | G | G | G | G | A | G | G | G | T | G | G | G | G | A | G | G | G | T | G | G | G | G | A | 3′ |
| 1-2-1 24 mer | 5′- | T | T | T | T | T | A | G | G | G | T | G | G | G | G | A | G | G | G | T | G | G | G | G | A | 3′ |
| 1-6-1 24 mer | 5′- | T | G | G | G | G | A | G | G | G | T | T | T | T | T | A | G | G | G | T | G | G | G | G | A | 3′ |
Results
The results of the DSC and CD experiments present a consistent picture of the structure and stability of the three model quadruplex-forming c-MYC promoter oligonucleotides. We found that the WT sequence folds into at least two unique structural quadruplexes with different thermal stabilities. We also observed that two mutant sequence oligonucleotides that are known to fold into unique 1:6:1 and 1:2:1 loop isomers exhibit a thermal stability similar to that of the two conformers present in WT solutions. Presumably, these two mutant sequences that fold into independent G-quadruplex structures adequately describe the mixture of folded conformations that exist in equilibrium for solutions containing the WT oligomer.
DSC thermal melting profiles for the WT 24-mer purine rich c-MYC promoter region oligonucleotide sequence are shown in Figs. 1 and 2. Fig. 1 shows the superimposed melting curves for the WT oligomer at two different potassium ion concentrations (30 mM [K+] and 130 mM [K+]). The WT thermograms at both salt concentrations are composed of a rather broad asymmetric peak. These broad transitions were deconvoluted into two two-state transitions needed to fit the thermogram within expected experimental error, as shown in Fig. 2. The two Tm-values for the two WT c-MYC 24 mer conformers are given in Table 1 for both ionic strengths. At 30 mM [K+], the lower melting transition of the WT has a Tm of 71.0°C and the higher melting transition has a Tm of 80.1°C. At 130 mM [K+], the lower melting transition of the WT has a Tm of 78.8°C and the higher melting transition has a Tm of 93.1°C.
Figure 1.
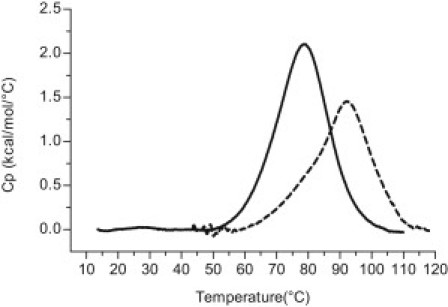
Raw excess heat capacity of the c-MYC WT P1 promoter 24-mer G-quadruplex at two different ionic strengths. The solid curve is the thermal denaturation of the WT 24-mer construct in 30 mM [K+] BPES buffer, whereas the dashed line is the thermal denaturation of the WT 24-mer construct in 130 mM [K+] BPES buffer at pH 7.0.
Figure 2.
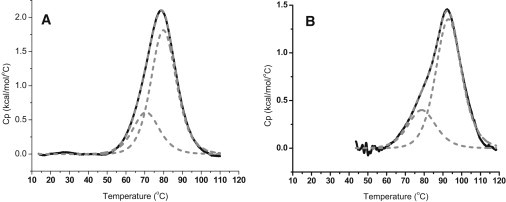
DSC thermograms for the thermal denaturation of the c-MYC WT P1 promoter 24-mer G-quadruplex in (a) 30 mM [K+] and (b) 130 mM [K+] BPES. In the lower ionic strength solution, the raw excess heat capacity has been deconvoluted into two “two-state processes.” The lower melting transition is attributed to the 1-6-1 isomer with a Tm of 72.4°C, and the higher melting conformer is attributed to the 1-2-1 isomer (Tm of 80.6°C). The raw excess heat capacity has been deconvoluted into two “two-state processes.” The lower melting conformer is attributed to the 1-6-1 isomer (Tm of 78.8°C), and the higher melting conformer is attributed to the 1-2-1 isomer (Tm of 93.1°C).
Table 1.
Tm-values for WT, 1-6-1, and 1-2-1 sequences at 30 mM [K+] and 130 mM [K+]
| 30 mM [K+] | ||||
|---|---|---|---|---|
| 30 mM [K+] BPES | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | Tm4 (°C) |
| WT 24 mer | 71.0 | 80.1 | – | – |
| 1:2:1 24 mer | 77.0 | – | – | – |
| 1:6:1 24 mer | 64.3 | – | – | – |
| WT 24 mer/TMPyP4 | 67.5 | 78.8 | 103.7 | 115∗ |
| 130 mM [K+] | ||
|---|---|---|
| 130 mM [K+] BPES | Tm1 (°C) | Tm2 (°C) |
| WT 24 mer | 78.8 | 93.1 |
| 1:2:1 24 mer | 89.5 | – |
| 1:6:1 24 mer | 77.9 | – |
Tm4 is estimated from Tm4 ≈ Tm2 + (ΔTm(3-1)).
At 130 mM [K+], both WT melting transitions occur at higher temperatures than observed for the same transitions in the 30 mM [K+] salt solution. The higher salt concentration appears to stabilize both of the overlapping two-state transitions as previously reported (1).
DSC thermal melting profiles for the 1:6:1 and 1:2:1 mutant 24-mer purine rich c-MYC promoter region oligonucleotide sequences at the higher potassium ion concentration (130 mM [K+]) are shown in Fig. 3. Experiments were run at both ionic strengths, but only the higher ionic strength (130 mM [K+]) experiments are shown here. The 130 mM [K+] data are obviously more physiologically relevant. The 1:6:1 and 1:2:1 thermograms are comprised of a single symmetric peak and have been fit for a single two-state transition as shown in Fig. 3. The Tm-values for the c-MYC mutant 1:2:1 and 1:6:1 mutant 24-mer quadruplexes are also shown in Table 2 for both ionic strengths. At 30 mM [K+], the single melting transition of the 1:6:1 mutant has a Tm of 64.3°C and the 1:2:1 mutant has a Tm of 77.0°C. Neither of these two melting temperatures corresponds very well with the two WT transitions at the lower potassium concentration. At 130 mM [K+], there is reasonable agreement between the two mutant transitions and the two WT transitions. The single melting transition of the 1:6:1 mutant has a Tm of 77.9°C (compared with 78.8°C for the WT) and the single melting transition of the 1:2:1 mutant has a Tm of 89.5°C (compared with 93.1°C for the WT). With an increase in ionic strength, an increase in the thermal stability of both the 1:2:1 and 1:6:1 mutant constructs was observed.
Figure 3.
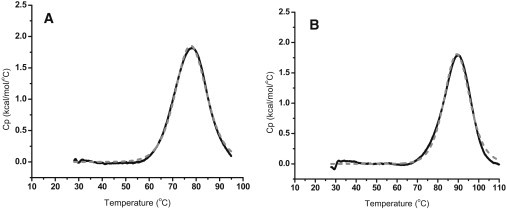
DSC thermograms for the thermal denaturation of the c-MYC mutant 24-mer G-quadruplex constructs in 130 mM [K+]. (a) Thermal denaturation of the 1-6-1 mutant 24-mer quadruplex. The raw excess heat capacity (dashed line) has been fit for a single “two-state process”(solid line). (b) Thermal denaturation of the 1-2-1 mutant 24-mer quadruplex. The raw excess heat capacity (dashed line) has been fit for a single two-state process (solid line).
Table 2.
Equilibrium constant and Gibbs free-energy change values for TMPyP4 binding to the WT, 1:6:1 mutant, and 1:2:1 mutant 24-mer c-MYC promoter quadruplex sequences at 30 mM [K+] and 130 mM [K+] salt concentrations
| 30 mM [K+] salt concentrations | ||||||
|---|---|---|---|---|---|---|
| 30 mM [K+] BPES | K1 (M−1) × 10−6 | ΔG1 | K2 (M−1) × 10−6 | ΔG2 | K3 (M−1) × 10−6 | ΔG3 |
| WT | 82 ± 1 | −10.6 | 3.0 ± 0.5 | −8.8 | – | – |
| 1-6-1 | 2020 ± 3 | −12.7 | 30.4 ± 1 | −10.2 | 1.3 ± 0.5 | −8.3 |
| 1-2-1 | 8320 ± 9 | −13.5 | 31.2 ± 1 | −10.2 | 1.9 ± 0.8 | −8.6 |
| 130 mM [K+] salt concentrations | ||||||
|---|---|---|---|---|---|---|
| 130 mM [K+] BPES | K1 (M−1) × 10−6 | ΔG1 | K2 (M−1) × 10−6 | ΔG2 | K3 (M−1) × 10−6 | ΔG3 |
| WT | 5 ± 0.5 | −9.1 | 0.2 ± 0.1 | −7.0 | – | – |
| 1-6-1 | 115 ± 3 | −11.3 | 10.8 ± 0.8 | −9.6 | 0.50 ± 0.08 | −7.6 |
| 1-2-1 | 86 ± 1 | −10.8 | 0.5 ± 0.1 | −7.7 | – | – |
The results of the DSC, ITC, UV-Vis, and CD titration experiments present a more complicated picture of the porphyrin-binding properties of the three model quadruplex-forming c-MYC promoter oligonucleotides. Fig. 4 shows the melting transitions for the c-MYC WT oligonucleotide in 30 mM [K+] after the addition of 1:10 moles of TMPyP4/WT DNA. In this case, the lower salt concentration was used to enlarge the DSC viewing window. At the higher ionic strength (130 mM [K+]), the melting transitions for both the free 1:2:1 conformer (Tm ≈ 93.1°C) and the TMPyP4 complexes (Tm estimated to be >115°C and >129°C for the 1:6:1 and 1:2:1 complexes, respectively) were too high to be accurately determined by DSC. In comparison with the melting curve for the WT-24 mer at 30 mM [K+], the following changes are observed: 1), both melting transitions are reduced in relative peak area (or enthalpy change); 2), a new thermal transition appears with a Tm ≥ 103°C, presumably due to the melting of the TMPyP4/G-quadruplex complex; and 3), the Tm for the 1:2:1/TMPyP4 complex is not observed but would be estimated to occur at a temperature above 115°C. With the appearance of the third melting transition (Tm ≈ 103°C), a decrease in the apparent enthalpy change (or peak area) for the 68°C melting transition is observed, which is consistent with the appearance of a new peak for the 1:6:1/TMPyP4 complex. We believe that the attenuation of the 78°C peak must reflect similar binding of TMPyP4 to the 1:2:1 conformer, and a new peak at 115°C would be expected to show approximately the same increase in stability for the 1:2:1 TMPyP4 complex.
Figure 4.
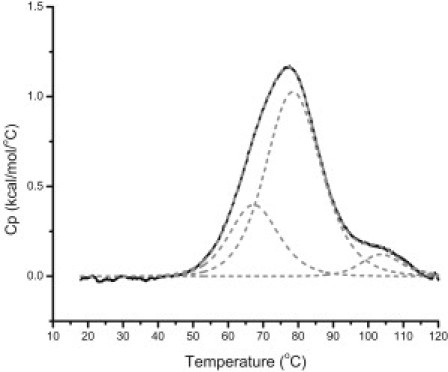
DSC thermogram for the thermal denaturation of the c-MYC WT 24-mer G-quadruplex with 1:10 mols of TMPyP4/WT DNA in 30 mM [K+]. The raw excess heat capacity has been deconvoluted into three independent two-state processes. The lower melting conformer is attributed to the 1-6-1 isomer, and the higher melting conformer is attributed to the 1-2-1 isomer. The highest melting transition, which appears with the addition of TMPyP4, is for the melting of the TMPyP4 stabilized G-quadruplex complex.
Fig. 5 shows data from a typical ITC titration for the addition of TMPyP4 to the 1:6:1 mutant 24-mer oligonucleotide. The figure shows the raw titration data, the integrated heat data, and the best-fit line for one of the three replicate titrations. The fit line shown in Fig. 5 is for a three-independent-sites model (1,27–30). The average thermodynamic parameters obtained for each set of replicate titrations are listed in Tables 2 (Ki and ΔGi values) and 3 (ΔHi and −TΔSi values) for either a two-site or three-site model as required for each different oligonucleotide and ionic strength. The ITC results obtained for both mutant oligonucleotides and the WT sequence quadruplex demonstrate a binding stoichiometry of 4:1 TMPyP4/DNA at saturation (1,27). The microcalorimetric integrated heat data for the titration of the c-MYC WT quadruplex are obviously complex but can be fit within experimental error for a binding model with at least two independent overlapping binding processes (exterior or end-binding and an “intercalative” binding mode). The mutant quadruplex titration data are more complex, requiring the use of a three-independent-binding-sites model to fit the thermograms for both the 1:6:1 and 1:2:1 mutants at the lower ionic strength and the 1:6:1 titration data at the higher ionic strength within expected experimental error. Presumably, the three-site independent overlapping binding model is consistent with one high-affinity end-binding site, two lower-affinity intercalation sites, and one still-lower-affinity end-binding site (1). The differentiation between TMPyP4 binding at the two ends of the two mutant sequences must be due to differences in folding of the terminal residues brought about by the numerous G→T substitutions, and may also be due to the large loop present in the 1:6:1 structure.
Figure 5.
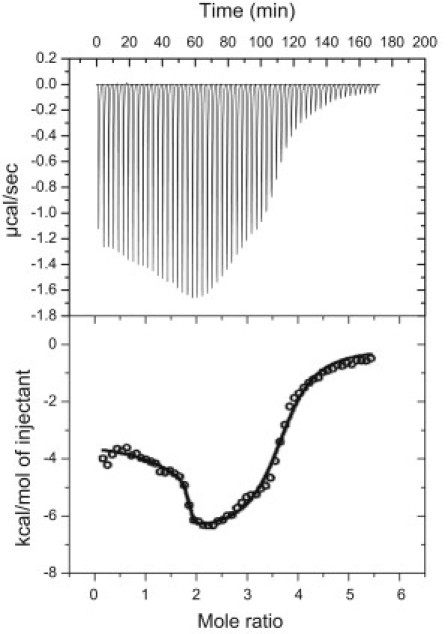
ITC data for the addition of TMPyP4 to the 1:6:1 c-MYC mutant construct at pH 7.0 in 130 mM [K+]. The upper panel shows the raw ITC power signal, and the lower panel shows the integrated heat data (○). The best-fit nonlinear regression line (—) is for the thermodynamic parameters obtained using a three-independent-sites model with three values for Ki, three values for ΔHi, and three values for ni.
Table 3.
ITC-derived thermodynamic parameters for TMPyP4 binding to the WT, 1:6:1 mutant, and 1:2:1 mutant 24-mer c-MYC promoter quadruplex sequences
| 30 mM [K+] salt concentration | ||||||
|---|---|---|---|---|---|---|
| 30 mM [K+] BPES | ΔH1 (kcal/mol) | −TΔS1 (kcal/mol) | ΔH2 (kcal/mol) | −TΔS2 (kcal/mol) | ΔH3 (kcal/mol) | −TΔS3 (kcal/mol) |
| WT | −1.7 (±0.1) | −9.0 | −11.2 (±0.2) | 2.3 | – | – |
| 1-6-1 | −6.0 (±0.2) | −6.6 | −11.4 (±0.8) | 1.2 | −16.8 (±1.3) | 8.4 |
| 1-2-1 | −6.0 (±0.1) | −7.5 | −10.1 (±0.8) | −2.8 | −16.3 (±0.6) | 7.7 |
| 130 mM [K+] salt concentration | ||||||
| 130 mM [K+] BPES | ΔH1 (kcal/mol) | −TΔS1 (kcal/mol) | ΔH2 (kcal/mol) | −TΔS2 (kcal/mol) | ΔH3 (kcal/mol) | −TΔS3 (kcal/mol) |
| WT | −0.7 (±0.1) | −8.4 | −8.2 (±1.7) | 1.3 | – | – |
| 1-6-1 | −3.5 (±0.2) | −8.0 | −8.6 (±1.7) | −1.0 | −5.0 (±0.9) | −2.7 |
| 1-2-1 | −3.5 (±0.2) | −7.3 | −7.6 (±0.4) | −0.2 | – | – |
Tables 2 and 3 list the Gibb's free-energy changes, binding constants, and the best-fit thermodynamic parameters for the binding of TMPyP4 to WT, 1:2:1 and 1:6:1 sequences obtained from the two- or three-independent-sites models. At the lower ionic strength (30 mM [K+]), the 1:2:1 conformer exhibits a slightly higher affinity for TMPyP4 for all three binding sites compared with the 1:6:1 conformer. Both the 1:2:1 and 1:6:1 mutant conformers exhibit higher (by 25–100 times) TMPyP4 affinity than the average affinity exhibited by the WT mixture of conformers. The binding of TMPyP4 to the 1:2:1 highest-affinity site (K1 = 8.3 × 109) is ∼200 times greater than the affinity for binding to the second and third 1:2:1 binding sites (K2= 3.1 × 107). The fourth and last 1:2:1 binding site exhibits the weakest affinity (K3 = 1.9 × 106). At the lower ionic strength, the WT DSC data, obtained at low mole ratios of TMPyP4/DNA (e.g., 1:10), are consistent with the ITC results in that TMPyP4 is shown to bind to both the 1:2:1 and 1:6:1 conformers with similar affinity.
At the higher ionic strength (130 mM [K+]), the 1:6:1 conformer exhibits a slightly higher affinity for TMPyP4 for all three binding sites compared with the 1:2:1 conformer. The binding of TMPyP4 to the 1:6:1 conformer highest-affinity site (K1= 2.9 × 108) is ∼27 times tighter than for the second and third binding sites (K2 = 1.1 × 107), which is ∼22 times greater than for binding to the fourth and last site (K3 = 4.9 × 105).
We performed UV-Vis titration experiments on the c-MYC WT sequence as well as the mutant 1:2:1 and 1:6:1 sequences at both ionic strengths to verify the binding stoichiometries calculated from the ITC experiments. The changes in absorbance were monitored after every injection. The corresponding job plot produced in these titrations produced break points at the mole ratios of 2:1 and 4:1 for TMPyP4/c-MYC DNA as well as an obvious end point at 4:1 moles of TMPyP4/c-MYC DNA for the 1:6:1 and 1:2:1 mutant oligonucleotide constructs (data not shown). These results are consistent with previously reported WT data (1). The titrations of 1:2:1 mutant, 1:6:1 mutant, and WT 24 mer oligonucleotides with TMPyP4 produced an obvious red shift in the maximum absorbance, indicating intercalation of the TMPyP4 into the quadruplex structure of each sequence. This is consistent with previously published results (1,27).
CD measurements of the WT, 1-2-1 mutant, and 1-6-1 mutant 24 mer quadruplex constructs at both ionic strengths demonstrated a spectral signature that was consistent with previously published intramolecular parallel stranded G-quadruplex DNA spectra (9). We performed the titrations up to 6:1 mols of TMPyP4/mol DNA to observe complete saturation of the oligonucleotide. The CD spectra for the 1:6:1 sequence at 130 mM [K+] alone, as well as solutions containing 0.5:1, 1:1, 2:1, 3:1, 4:1, 5:1, and 6:1 moles of TMPyP4/mole of the 1:6:1 oligomer are shown in Fig. 6. The CD spectra for the 1:6:1G-quadruplex TMPyP4 complex exhibits an attenuation of the 265 nm ellipticity and an increase in ellipticity at 290 nm with increasing TMPyP4. In contrast, the CD titration data for the WT and 1:2:1 mutant G-quadruplexes (data not shown) are completely insensitive to the addition of TMPyP4 (1). The WT and 1:2:1 G-quadruplexes apparently remain in the intramolecular parallel folded configuration, even after the addition of saturating amounts of TMPyP4, whereas the 1:6:1 G-quadruplex must undergo some refolding to include some antiparallel structure upon addition of TMPyP4.
Figure 6.
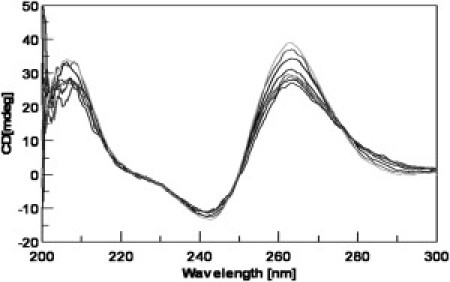
CD spectra of the c-MYC P1 1-6-1 24-mer G-quadruplex construct in 130 mM [K+] BPES titrated with TMPyP4. Typical CD spectra are shown for the 1-6-1 mutant alone (light gray), and the consecutive spectra from top to bottom are for the complex of DNA/TMPyP4 with mole ratios of 1:0, 1:.5, 1:1, 1:2, 1:3, 1:4 1:5, and 1:6. The CD spectrum for the G-quadruplex sequence at pH 7.0 shows the classical intramolecular parallel-stranded quadruplex with a characteristic maximum in molar ellipticity at ∼264 nm and a characteristic minimum in molar ellipticity at ∼242 nm.
Bound TMPyP4 did not exhibit an induced CD signal in any of the c-MYC G-quadruplex complexes studied here (data not shown). Under similar experimental conditions, we previously observed that TMPyP4 did not exhibit a induced CD signal but did continue to exhibit a characteristic absorbance spectrum with a red shift on binding (1). The TMPyP4 absorbance increases according to the free drug extinction coefficient once the oligonucleotide has been saturated with TMPyP4 (4:1 moles of TMPyP4/oligonucleotide).
Discussion
We have demonstrated that the c-MYC P1 promoter is a complex system comprised of at least two unique quadruplex structures in equilibrium with each other and the random coil. Our data are consistent with studies performed by Phan et al. (8) in which two mutant sequences were shown to fold into unique G-quadruplex structures in [K+] solution. Phan et al. showed that the two quadruplex structures are composed of a core of three stacked G-tetrads formed by four G-stretches, forming three parallel G-tetrads, and three double-chain-reversal loops bridging the three planar G-tetrads. The mutant quadruplexes contain either one, two, and one, or one, six, and one unstructured bases in the connecting lateral loops. Using NMR, Phan et al. (8) found the melting temperature for the lower melting mutant sequence at 90 mM [K+] to be ∼75°C, which is comparable to the temperature of our 1:6:1 mutant at 130 mM (i.e., 77.9°C). However, at the 90 mM ionic strength, Phan et al. were unable to determine the melting temperature of the higher melting mutant, since the melting temperature was above 80° and could not be measured within the limits of the NMR instrument. Using DSC, we observed a melting temperature for the higher melting conformer at 130 mM [K+] of 89.5°C. At a very low ionic strength (9 mM [K+]), Phan et al. (8) found an ∼16°C difference in melting temperature between the lower melting (1:6:1) and higher melting (1:2:1) mutants. Their reported 16°C difference between the melting temperatures of the lower melting and higher melting constructs is similar to our DSC ΔTm results of 12.7°C and 11.6°C for the 1:6:1 and 1:2:1 mutants at 30 mM [K+] and 130 mM [K+], respectively.
DSC has been shown to be a valuable technique for unambiguously determining the minimum number of unique conformations (with different thermal stabilities) that exist in equilibrium in dilute solutions of single-stranded polypurine sequences capable of forming G-quadruplexes (8). Seenisamy et al. (9) proposed four folding topologies for the c-MYC WT 24-mer polypurine sequence, with the different conformers having either one or two bases in the connecting loops, and the two most favorable topologies having two different sets of four G-tracks. Phan et al. (8) stated that out of the six available guanine runs (with three or more consecutive guanines) in the WT sequence, there are five different possibilities for formation of a stabile G-quadruplex structure. They examined two of the five folding topologies using NMR and mutant sequences restricted to form quadruplexes with either a 1:2:1 or 1:6:1 loop isomer structure. Our data, and in particular our DSC results, support the hypothesis that only two (of the possible five) folded isomers are needed to accurately describe the solution species distribution of c-MYC G-quadruplexes in equilibrium. The Tm for the 1:6:1 mutant sequence at both ionic strengths is similar to the Tm exhibited by the lower melting conformer in the WT sequence at the two ionic strengths. The Tm for the 1:2:1 mutant sequence at both ionic strengths is similar to the Tm exhibited by the higher melting WT quadruplex species at the same ionic strengths. We have clearly shown that the c-MYC WT 24-mer sequence exists as a mixture of at least two unique G-quadruplex structures. There may actually be additional quadruplex species in c-MYC oligonucleotide solutions; however, any additional species would need to have a melting profile very similar to that of either the 1:6:1 or 1:2:1 mutant sequence conformers.
The melting temperatures for all of the higher-order structured oligonucleotides (i.e., the c-MYC WT and loop isomer mutant sequences) increased with increasing ionic strength ([K+]). This is an expected result for any self-associated, intra- or intermolecular species. The higher ionic strength (or, in this case, the higher counter-cation concentration) attenuates the repulsive forces between the negatively charged phosphodiester groups. Phan et al. (8) observed similar trends in thermal stability for the WT and 1:2:1 and 1:6:1 mutant constructs. It is well known that in addition to a generic salt or ionic strength charge screening effect, the formation of these parallel quadruplex structures is driven somewhat by a specific ion effect. The quadruplex structures for these sequences are predicted to be different in K+- versus Na+-containing solutions. As we previously reported, the salt dependence of c-MYC quadruplex stability and salt effects on TMPyP4 binding are a combination of ionic strength and specific ion binding effects on the quadruplex structure in solution (1). The Tm can be predicted as a function of the potassium ion activity in the buffer solution (31), and, as shown here, it is also highly dependent on the number of unstructured bases in the quadruplex core structure. The least stable conformer at both ionic strengths examined here is the 1:6:1 conformer. From the DSC melting profiles, it can be assumed that the number of bases in the loops directly impacts the Tm of the G-quadruplex, since the Tm decreases as the number of unstructured bases in the lateral or end loops increases. At the lower ionic strength, the larger six-base loop of the 1:6:1 mutant must be more rigid and may not be flexible enough to participate in the end binding of TMPyP4. We propose that the increased affinity for TMPyP4 observed for the 1:6:1 mutant at the higher ionic strength reflects an increased ability of the loop bases to wrap around and facilitate the binding of TMPyP4. The two-base mid loop of the 1:2:1 conformer is too small to participate in TMPyP4 binding at either ionic strength.
After the addition of 1:10 mole of TMPyP4 per mole of WT DNA in the lower ionic strength solution (30 mM [K+]), both the lower and higher temperature melting transitions are attenuated. Concomitantly, a third melting profile with a fraction of the missing enthalpy appears with a Tm of ∼103°C. The new highest melting transition is assumed to result from the melting of the TMPyP4/1:6:1 complex. The melting of the TMPyP4/1:2:1 complex, inferred from the attenuated area for the 79°C peak, would be predicted to occur with a Tm above 115°C. This is consistent with the ITC-measured TMPyP4 affinities for both the 1:6:1 and 1:2:1 mutant sequences. Although the ΔTm observed for the 1:6:1 complex (12.7°C) is consistent with a high binding affinity, the uncertainty in determining an accurate Tm for melting of either the TMPyP4/1:6:1 or TMPyP4/1:2:1 complexes precludes accurate DSC determination of K for the formation of these ligand/DNA complexes.
The saturation stoichiometry for the binding of TMPyP4 to both the 1:6:1 and 1:2:1 mutant constructs is 4:1 mole (TMPyP4/DNA), a result that is consistent with previous TMPyP4/quadruplex DNA binding studies (1). Upon addition of TMPyP4, the CD and UV-Vis results demonstrated that after saturation, the c-MYC 24-mer quadruplex structure was still present and relatively unchanged. ITC titrations were performed on the three c-MYC sequences (WT, 1:2:1, and 1:6:1) with TMPyP4. The ITC results demonstrated that at the low ionic strength (30 mM [K+]), the WT construct exhibited only two modes of binding, whereas the 1:6:1and 1:2:1 constructs exhibited three modes of binding. At the higher ionic strength (130 mM [K+]), the 1:6:1 construct was the only one that exhibited three modes of binding; the WT and 1:2:1 construct exhibited only two binding modes.
The energetic profile for the first TMPyP4 binding mode is indicative of an entropy-driven process. In the case of hydrophobic ligands binding in the DNA minor groove, water solvating the free ligand and structured water from the minor groove are expelled, resulting in a large positive change in entropy (32–36). We speculate here that the entropy-driven energetics for mode 1 binding, which is typical for the binding of a hydrophobic ligand to the exterior of duplex DNA, can be used as a model for the exterior or end-binding interactions of the TMPyP4 ligand to the top or bottom of the G-quadruplex structure. One subtle difference between exterior binding to the G-quadruplex and minor groove binding to duplex DNA is that the water on the exterior of the G-quadruplex is probably less organized than groove or spine water in duplex DNA. The energetic profile for the second TMPyP4 binding mode is indicative of an enthalpy-driven process. In the case of the insertion of planar aromatic ligands between the stacked bases in duplex DNA, the typical exothermic enthalpy change is largely the result of increased π-π stacking interactions between the DNA bases and the intercalated aromatic ligand (32–36). We speculate that the enthalpy-driven energetics for mode 2 binding, which is typical for the intercalation of a planar aromatic ligand into duplex DNA, can be used as a model for the stacking of the TMPyP4 ligand between G-tetrads in the G-quadruplex structure. A subtle difference would be that in duplex intercalation, there are additional consequences of intercalation, such as some unwinding of the DNA duplex.
Although the intercalation binding mode remains hypothetical, there continue to be reports that intercalation can occur (24,37–40). Gavathiotis et al. (37) reported an NMR structure (PDB 1NZM) with a spacing of 6.9 Å between a G-tetrad and a plane of four adenines in a parallel-stranded DNA quadruplex, d(TTAGGGT)4, containing the human telomere repeat sequence and intercalated quinoacridinium ligands. Keating and Szalai (24) reported electron paramagnetic resonance data consistent with the intercalation of CuTMPyP4 into a d(T4G8T4)4 G-quadruplex. Hounsou et al. (38) reported an NMR structure (PDB 2JWQ) with a spacing of 5.8 Å between a G-tetrad and a plane of four adenines, with quinacridine-based ligands (MMQs) intercalated between two G- and A-planes. Zhang et al. (39) reported an NMR structure (PDB 2KKA) for a two G-tetrad intramolecular G-quadruplex with a tetrad spacing of 4.7 Å. A molecular-dynamics simulation study by Cavallari et al. (40) revealed that TMPyP can stack with G-tetrads in the absence of interplane cations. They reported TMPyP G-tetrad stacking distances of 4.3–4.7 Å and noted that TMPyP intercalation depends on the length of the quadruplex, the stoichiometric ratio, and the edge termination motif. Although none of these studies directly demonstrated TMPyP4 intercalation between G-tetrads in an intramolecular G-quadruplex, we believe that the possibility of intercalation cannot be excluded at this time. Our own previous modeling study demonstrated the possibility of a saturated c-MYC TMPyP4/G-quadruplex having two end stacked ligands and two intercalated ligands (41). In that case, the separation of the internal G-Tetrads was 6.2 Å, which would be large enough to accommodate the planar TMPyP4 ligand without a significant increase in tetrad spacing.
The loss of the third binding mode of the 1:2:1 at the higher ionic strength is evidence that the [K+] concentration does not affect the core of the G-quadruplex structure, as shown by the CD results in Fig. 6, but does affect the unpaired bases that are not involved in G-quadruplex formation. Mode 3 binding, which is principally observed at low ionic strength, looks like a combination of an enthalpically favorable stacking interaction and an entropically unfavorable restructuring of one or more G-quadruplex end loops. At the higher ionic strength, the end loops are more flexible and less restructuring is needed to accommodate ligand binding to the hindered end of the 1:6:1 G-quadruplex. Differences in the number of binding modes exhibited by the 1:6:1 and 1:2:1 mutant quadruplexes in higher-ionic-strength solution must be due to the number and structure of the bases in the loop and tail regions. The 1:2:1 construct exhibits only end-binding modes that do not involve loop bases (with both ends behaving in the same manner) and intercalation. At the higher ionic strength, the third binding mode for the 1:6:1 construct is assigned to an end-binding process that involves restructuring of the large six-base loop. This binding mode exhibits a more favorable enthalpy change and a less favorable entropy change in comparison with mode 1 binding. We attribute these differences to the coupling of the favorable enthalpy effect of stacking loop bases over the ligand on the hindered end of the 1:6:1 construct with the unfavorable entropy for restructuring the loop bases.
A more problematic result is that the average TMPyP4 affinity for the c-MYC WT 24-mer sequence is significantly weaker than that for either the 1:2:1 or 1:6:1 construct. This observation holds for both ionic strengths and both the end and intercalation binding modes. When we compare it with the highest-affinity binding site at the lower ionic strength (30 mM [K+]), the TMPyP4 affinity follows the trend K1,WT << K1,1:6:1 < K1,1:2:1, with K1,1:2:1 being ∼1000 times greater than K1,WT. Both the 1:6:1 and 1:2:1 mutant constructs exhibit a very unfavorable entropy change for the third binding mode that is not seen for the WT (presumably an equilibrium mixture of the 1:6:1 and 1:2:1 species). When we compare it with the highest-affinity binding site at the higher ionic strength (130 mM [K+]), the TMPyP4 affinity follows the trend K1,WT << K1,1:2:1 < K1,1:6:1, with K1,1:6:1 being ∼60 times greater than K1,WT. Again, the thermodynamic profile for the TMPyP4-WT interactions (presumably an equilibrium mixture of the 1:6:1 and 1:2:1 species) is not the simple average of the two mutant construct interactions.
One plausible explanation is that WT solutions contain a much richer mixture of conformations in dynamic equilibrium and are not well modeled as a mixture of the two constrained (1:6:1 and 1:2:1) species on which we focused in this study. The energetic and dynamic barriers for the folding/unfolding/refolding processes could potentially explain the differences in the thermodynamics for binding TMPyP4 to these various structures.
Conclusions
The c-MYC WT 24-mer guanine-rich sequence adopts at least two unique conformations at both ionic strengths (30 mM and 130 mM [K+] BPES). It is obvious that noncore bases affect both the quadruplex binding and stability characteristics. We have demonstrated that DSC melting profiles can be used to evaluate and/or deconvolute the different structural species that might exist in equilibrium for a folded oligonucleotide. We have also shown that the lower melting species in an equilibrium mixture of WT sequence quadruplexes appears to be the 1:6:1 loop isomer described by Phan et al. (8), and that the higher melting species appears to be the 1:2:1 loop isomer. The strength of this study lies in part in the correlation and/or agreement between the DSC and ITC data. Future attempts to search for compounds with a high selectivity and affinity for the c-MYC promoter region quadruplex should take advantage of the mid-loop involvement in the high-affinity binding process. This is especially important in light of the fact that studies of drug activity should focus on interactions at low mole ratios, certainly below saturating levels. The results presented here have solidified our understanding of the c-MYC quadruplex equilibria and may potentially lead to the design of new and better G-quadruplex selective agents.
Supporting Material
References
- 1.Freyer M.W., Buscaglia R., Lewis E.A. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophys. J. 2007;92:2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dexheimer T.S., Sun D., Hurley L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed J.E., Arnal A.A., Vilar R. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel(II) complexes. J. Am. Chem. Soc. 2006;128:5992–5993. doi: 10.1021/ja058509n. [DOI] [PubMed] [Google Scholar]
- 4.Kaushik M., Bansal A., Kukreti S. Possibility of an antiparallel (tetramer) quadruplex exhibited by the double repeat of the human telomere. Biochemistry. 2007;46:7119–7131. doi: 10.1021/bi0621009. [DOI] [PubMed] [Google Scholar]
- 5.Kumar P., Verma A., Chowdhury S. Tetraplex DNA transitions within the human c-myc promoter detected by multivariate curve resolution of fluorescence resonance energy transfer. Biochemistry. 2005;44:16426–16434. doi: 10.1021/bi051452x. [DOI] [PubMed] [Google Scholar]
- 6.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley L.H. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem. Soc. Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 8.Phan A.T., Modi Y.S., Patel D.J. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seenisamy J., Rezler E.M., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 10.Grand C.L., Powell T.J., Hurley L.H. Mutations in the G-quadruplex silencer element and their relationship to c-MYC overexpression, NM23 repression, and therapeutic rescue. Proc. Natl. Acad. Sci. USA. 2004;101:6140–6145. doi: 10.1073/pnas.0400460101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Li W., Miyoshi D., Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 12.Siddiqui-Jain A., Grand C.L., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelengaris S., Rudolph B., Littlewood T. Action of Myc in vivo—proliferation and apoptosis. Curr. Opin. Genet. Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 14.Kim M.Y., Duan W., Hurley L.H. Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase II and G-quadruplexes. J. Med. Chem. 2003;46:571–583. doi: 10.1021/jm0203377. [DOI] [PubMed] [Google Scholar]
- 15.Rezler E.M., Bearss D.J., Hurley L.H. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 16.Halder K., Mathur V., Chowdhury S. Quadruplex-duplex competition in the nuclease hypersensitive element of human c-myc promoter: C to T mutation in C-rich strand enhances duplex association. Biochem. Biophys. Res. Commun. 2005;327:49–56. doi: 10.1016/j.bbrc.2004.11.137. [DOI] [PubMed] [Google Scholar]
- 17.Risitano A., Fox K.R. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 18.Haider S.M., Parkinson G.N., Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 19.Wheelhouse R.T., Sun D., Hurley L.H. Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J. Am. Chem. Soc. 1998;120:3261–3262. [Google Scholar]
- 20.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 21.Chen L., Cai L., Rich A. Crystal structure of a four-stranded intercalated DNA: d(C4) Biochemistry. 1994;33:13540–13546. doi: 10.1021/bi00250a005. [DOI] [PubMed] [Google Scholar]
- 22.Anantha N.V., Azam M., Sheardy R.D. Porphyrin binding to quadrupled T4G4. Biochemistry. 1998;37:2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- 23.Sehlstedt U., Kim S.K., Dabrowiak J.C. Interaction of cationic porphyrins with DNA. Biochemistry. 1994;33:417–426. doi: 10.1021/bi00168a005. [DOI] [PubMed] [Google Scholar]
- 24.Keating L.R., Szalai V.A. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry. 2004;43:15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 25.Seenisamy J., Bashyam S., Hurley L.H. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J. Am. Chem. Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted in proof.
- 27.Nagesh N., Buscaglia R., Lewis E.A. Studies on the site and mode of TMPyP4 interactions with Bcl-2 promoter sequence G-quadruplexes. Biophys. J. 2010;98:2628–2633. doi: 10.1016/j.bpj.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freyer M.W., Buscaglia R., Lewis E.A. Binding of netropsin and 4,6-diamidino-2-phenylindole to an A2T2 DNA hairpin: a comparison of biophysical techniques. Anal. Biochem. 2006;355:259–266. doi: 10.1016/j.ab.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 29.Freyer M.W., Lewis E.A. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 30.Dettler J.M., Buscaglia R., Lewis E.A. Biophysical characterization of an ensemble of intramolecular i-motifs formed by the human c-MYC NHE III1 P1 promoter mutant sequence. Biophys. J. 2010;99:561–567. doi: 10.1016/j.bpj.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrus A., Chen D., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 32.Chaires J.B. Analysis and interpretation of ligand-DNA binding isotherms. Methods Enzymol. 2001;340:3–22. doi: 10.1016/s0076-6879(01)40415-0. [DOI] [PubMed] [Google Scholar]
- 33.Suh D., Chaires J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995;3:723–728. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]
- 34.Ren J., Chaires J.B. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 35.Hyun K.M., Choi S.D., Kim S.K. Can energy transfer be an indicator for DNA intercalation? Biochim. Biophys. Acta. 1997;1334:312–316. doi: 10.1016/s0304-4165(96)00111-0. [DOI] [PubMed] [Google Scholar]
- 36.Chaires J.B. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Gavathiotis E., Heald R.A., Searle M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Hounsou C., Guittat L., Teulade-Fichou M.P. G-quadruplex recognition by quinacridines: a SAR, NMR, and biological study. ChemMedChem. 2007;2:655–666. doi: 10.1002/cmdc.200600286. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Dai J., Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavallari M., Garbesi A., Di Felice R. Porphyrin intercalation in G4-DNA quadruplexes by molecular dynamics simulations. J. Phys. Chem. B. 2009;113:13152–13160. doi: 10.1021/jp9039226. [DOI] [PubMed] [Google Scholar]
- 41.Cashman D.J., Buscaglia R., Lewis E.A. Molecular modeling and biophysical analysis of the c-MYC NHE-III1 silencer element. J. Mol. Model. 2008;14:93–101. doi: 10.1007/s00894-007-0254-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


