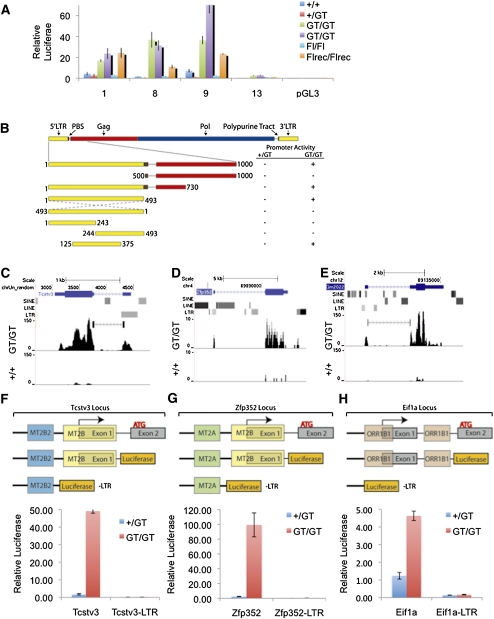
Figure 4.
Endogenous retroviral LTRs are activated in Kdm1a mutant ES cells. (A) Four unique MERVL LTR-Gag reporter luciferase plasmids (1, 8, 9, and 13) were cotransfected with CMV βgal into Kdm1a wild-type or mutant ES cells as indicated, and relative luciferase levels were measured 24 h later. Error bars represent SEM. (B) Schematic of MERVL retrovirus and truncation mutants generated and tested for promoter activity in Kdm1a wild-type and mutant cells. Luciferase activity (at least 20-fold over background) is indicated with a plus sign (+). (C–E) Raw mRNA-seq reads from wild-type (+/+) and Kdm1a GT/GT ES cells were aligned to the mouse genome and displayed on the UCSC genome browser at the Tcstv3 (C), Zfp352 (D), and Eif1a (Gm2022) (E) genes with RepeatMasker function enabled to visualize the position of LTR elements. (C,E) Detected junctions are also displayed for Tcstv3 and Gm2022. (F–H) The promoter-proximal sequences of Tcstv3 (F), Zfp352 (G), and Eif1a (Gm2022) (H), as well as deletion constructs (−LTR), as schematized above, were cloned upstream of a promoterless luciferase reporter gene and cotransfected with a CMV βgal reporter into Kdm1a +/GT or GT/GT ES cells. Relative luciferase activity was measured 24 h later and plotted, with error bars representing SEM.